Figures & data
Table 1 List of primers used for DPYD exon sequencing
Figure 1 Sequence electropherogram of the 2242+1G>T variant. Blue, dCTP; green, dATP; black, dGTP; red, dTTP.
Abbreviations: dATP, deoxyadenosine triphosphate; dCTP, deoxycytidine triphosphate; dGTP, deoxyguanosine triphosphate; DPYD, dihydropyrimidine dehydrogenase; dTTP, deoxythymidine triphosphate.
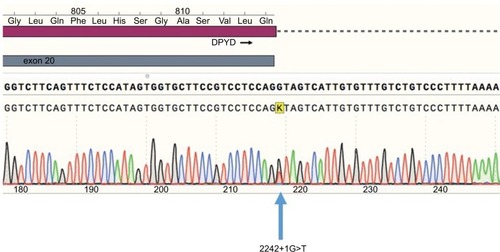
Figure 2 Exon skipping sequence.
Notes: Left: Electrophoresis of amplified cDNA from peripheral blood mononuclear cells of a patient carrying the 2242+1G>T variant. Right, upper: Graphic representation of skipping of exon 19. Right, lower: Sequence electropherogram using the primer Ex18F. A double sequence is observed for 85 nucleotides corresponding to overlapping of exons 19 and 20.
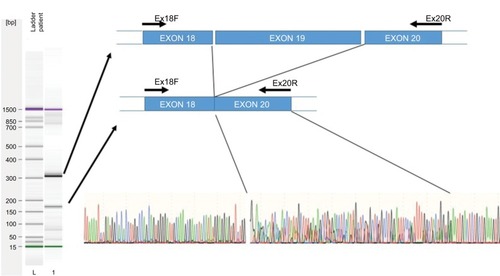
Figure 3 Structures of wild-type DPYD and truncated DPYD.
Notes: The interpretation of mutation effect and the molecular modeling were performed by using Deep View Swiss-PDB viewer and Tasser. (A) Modeling of wild-type DPYD; (B) modeling of truncated DPYD.
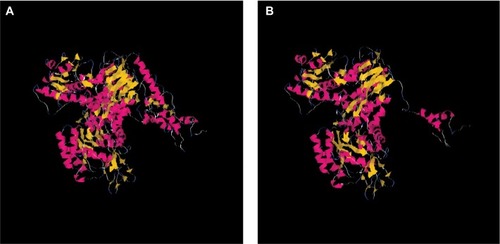
Table 2 DPYD phenotyping results
