Figures & data
Table 1 The relationship between miR-204 expression and clinicopathological features in PTC patients from TCGA data set
Figure 1 Heatmap of the 181 dysregulated miRNAs.
Notes: Hierarchical analysis of the 181 dysregulated miRNAs based on the comparison of their expression values in PTC and normal tissues. Data have been deposited in GEO (accession code GSE113629).
Abbreviations: GEO, Gene Expression Omnibus; miRNAs, microRNAs; PTC, papillary thyroid carcinoma; T, tumor tissues; N, normal tissues.
Figure 2 Downregulated miR-204 expression in PTC tissues and cell lines.
Notes: (A) miR-204 expression levels are downregulated in human PTC tissues compared to those in paired adjacent normal tissues. (B) miR-204 expression levels are downregulated in human PTC cell lines compare to that in normal cell line. *P<0.05.
Abbreviation: PTC, papillary thyroid carcinoma.
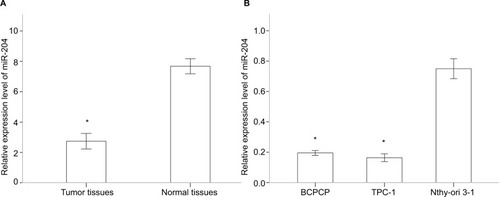
Table 2 Comparison of methylation levels in CpG sites between PTC and adjacent normal tissues
Figure 3 Promoter DNA methylation analysis of miR-204.
Notes: (A) Schematic representation of the location of miR-204 in chromosome 9. miR-204 is located within the introns of TRPM3 (located at 9q21). (B) CpG island prediction and primer design of the miR-204 promoter region. Two CpG islands around the miR-204 promoter were identified using MethPrimer. Regions 9 and 15 that completely cover the two CpG islands were selected for the amplification. Each numbered region represents a region that can be amplified.
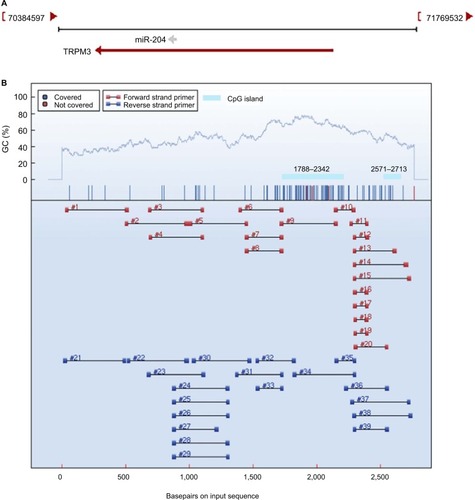
Figure 4 Promoter DNA methylation analysis of miR-204 with TCGA data.
Notes: (A) Correlation analysis between miR-204 expression levels and promoter methylation levels. The expression levels of miR-204 in tumor tissues were inversely correlated with the promoter DNA methylation rates. (B) Methylation levels of CpG sites from TCGA data. Three of the four CpG sites were hypermethylated in tumor tissues compared to normal tissues. One CpG site was hypomethylated (cg14223966). (C and D) qRT-PCR demonstrated increased expression of miR-204 in PTC cells after treatment with 5-Aza-dye for 48 and 98 hours compared to mock-treated cells. *P<0.05.
Abbreviations: 5-Aza, 5-aza-2′-deoxycytidine; PTC, papillary thyroid carcinoma; qRT-PCR, quantitative real-time PCR.
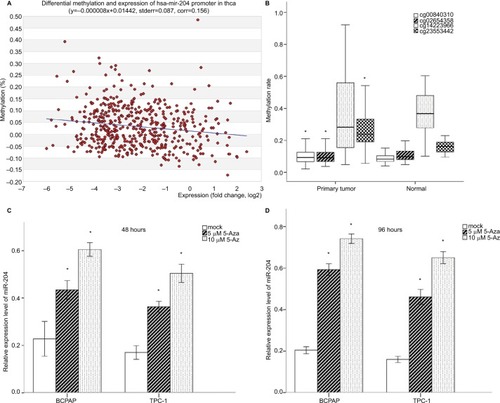
Figure 5 miR-204 promoter methylation rates in multiple cancers.
Notes: In 13 types of cancers, higher methylation rates were observed in cancer tissues than in normal tissues. *P<0.05, **P<0.005.
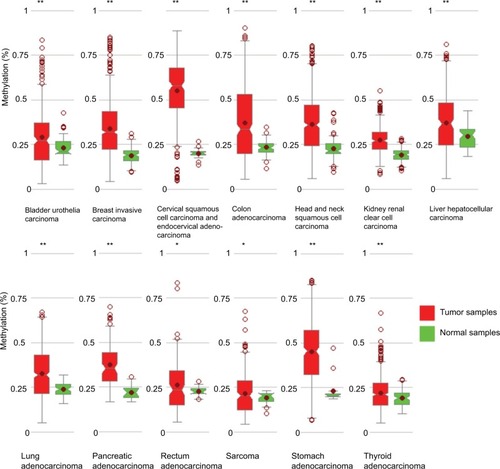
Figure 6 GO and KEGG enrichment analyses of miR-204-related DEGs.
Notes: (A) Significantly enriched GO terms of DEGs in PTC based on their functions. GO term classifications include biological process, cellular component, and molecular function. (B) Enriched KEGG pathways of miR-204-related DEGs. KEGG terms, gene numbers, P-values, and fold enrichment scores are presented in the pathway enrichment analysis. (C) Downregulation of miR-204-related pathway enrichment analysis by GSEA. The top enriched pathways showed significant enrichment in PTC tissues with lower miR-204 expression levels compared to tissues with higher miR-204 expression levels.
Abbreviations: DEG, differentially expressed gene; GO, gene ontology; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; PTC, papillary thyroid carcinoma.
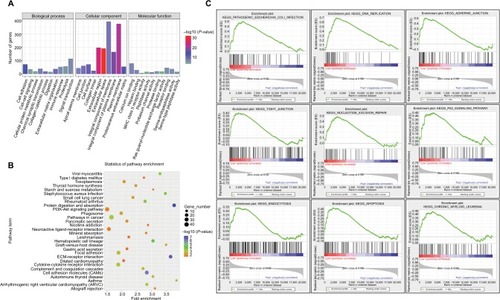
Figure S1 (A) Schematic representation of the chromatin patterns of miR-204. miR-204 is located within the introns of TRPM3 at 9q21. The genomic structure of the miR-204 and TRPM3 gene (currently known transcripts), the location of neighboring genes, and CpG islands are presented in each line. All data are obtained from the UCSC genome browser. (B) The genomic sequence of TRPM3 DNA promoter region. The promoter region was predicted from the genomic sequence of TRPM3 for the 2,000 upstream and 1,000 downstream sequences around its first exon (transcriptional start site). All of the CpG sites have been highlighted. CpG sites in red color are identified CpG sites of the gene chip data from TCGA.
Abbreviations: TCGA, the Cancer Genome Atlas; UCSC, University of California Santa Cruz.

