Figures & data
Figure 1 miR-4458 inhibited cell proliferation in NSCLC cells.
Notes: (A) Relative expression level of miR-4458 in four NSCLC cell lines (A549, H1299, HCC827, and PC9) and one normal lung cell (HBE). (B) The expression of miR-4458 after 48 hours of transfection with NC, mimics, inhibitor NC, and inhibitor by qRT-PCR. (C) The rate of cell growth was measured by Edu assay. (D) CCK-8 assay confirmed that miR-4458 inhibited cell proliferation in NSCLC cells. (E) The ability of colony formation was reduced with the overexpression of miR-4458 and enhanced with knockdown of miR-4458. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CCK-8, cell-counting kit-8; Edu, 5-ethynyl-2′-deoxyuridine; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
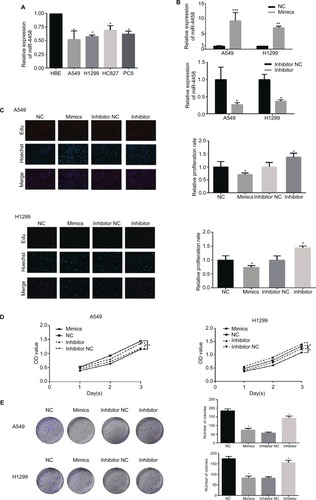
Figure 2 miR-4458 inhibited migration and the EMT process in NSCLC cells
Notes: (A) The results of transwell assay showed that miR-4458 reduced migration ability in NSCLC cells. (B) The results of wound-healing assay showed that miR-4458 inhibited migration ability. (C) Western blot analysis indicated that overexpression of miR-4458 upregulated EMT biomarker E-cadherin and reduced EMT biomarkers N-cadherin and Snail in NSCLC cells. Relatively, E-cadherin was downregulated and N-cadherin and Snail were enhanced with miR-4458 knockdown. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
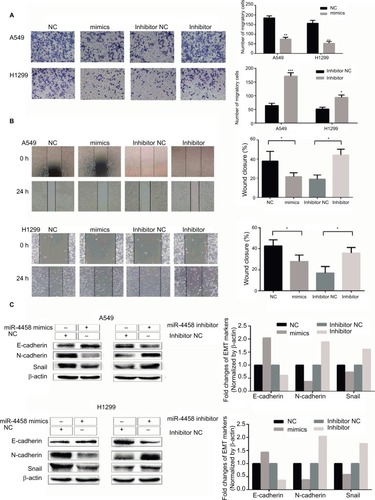
Figure 3 miR-4458 inhibited migration and the EMT process via PI3K/AKT pathway in NSCLC cells.
Notes: (A) The protein levels of p-AKT and AKT were evaluated by Western blot assay. (B) The results of transwell assay with miR-4458 knockdown were abolished by a PI3K inhibitor (LY294002). (C) The protein levels of E-cadherin, N-cadherin, Snail, p-AKT, and AKT were evaluated by the exposure of LY294002. **P<0.01.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NSCLC, non-small-cell lung cancer.
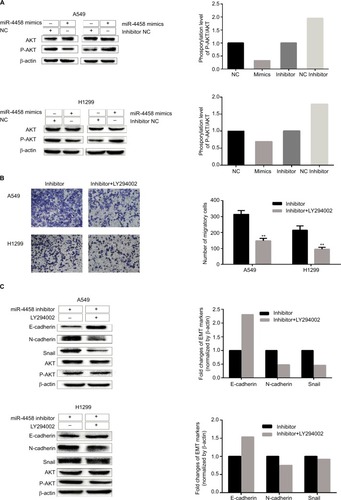
Figure 4 Bioinformatics analysis of the targets of miR-4458.
Notes: (A) Venn chart of common target genes from three publicly available databases including TargetScan, TarBase, and miRwalk. (B) GO analysis included biological processes, cellular component, molecular function, and pathway enrichment analysis for 176 common targets. (C) Venn chart of 8 more possible targets from 176 common target genes and TCGA database. (D) The expression of HMGA1 between LUAD and normal group in GSE32863 and GSE10072 datasets. (E) Analysis of the OS about HMGA1 from Kaplan–Meier plotter database. (F) The expression of HMGA1 in lung cancer cell lines from CCLE database. (G) The expression of HMGA1 between four NSCLC cell lines (A549, H1299, HCC827, and PC9) and one normal lung cell (HBE) by qRT-PCR. *P<0.05, **P<0.01.
Abbreviations: GO, Gene Ontology; miR-4458, microRNA-4458; NC, negative control; OS, overall survival.
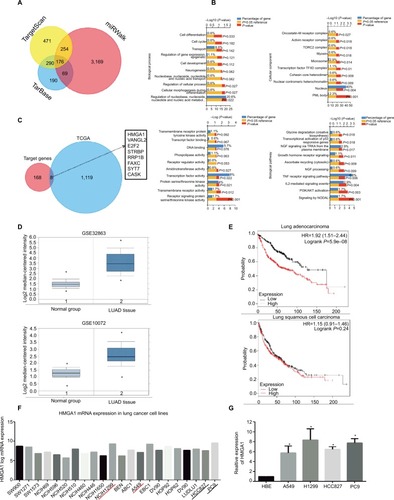
Figure 5 HMGA1 enhanced migration and the EMT process in NSCLC cells.
Notes: (A) The results of transwell assay showed that downregulation of HMGA1 reduced migration capacity in NSCLC cells. (B) The results of wound-healing assay showed that downregulation of HMGA1 inhibited migration capacity in NSCLC cells. (C) Western blot assay indicated that EMT biomarker E-cadherin was upregulated and N-cadherin and Snail were downregulated after knockdown of HMGA1. *P<0.05, **P<0.01.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
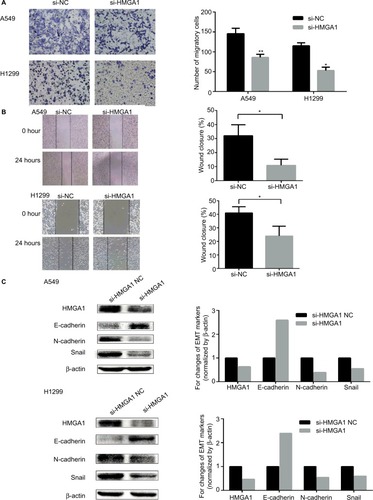
Figure 6 HMGA1 was a direct target of miR-4458.
Notes: (A) The predicted binding site of miR-4458 in the 3′-UTR of HMGA1 from miRNA website. (B) The analysis of luciferase activity after different combinations of miR-4458, miR-NC, h-HMGA1-WT, and h-HMGA1-MUT. (C) The expression of HMGA1 in NSCLC cells after 48 hours of transfection with miR-4458 NC, mimics, inhibitor NC, and inhibitor. (D) Western blot analysis of HMGA1 in NSCLC cells after 48 hours of transfection with miR-4458 NC, mimics, inhibitor NC, and inhibitor. *P<0.05, **P<0.01.
Abbreviations: miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
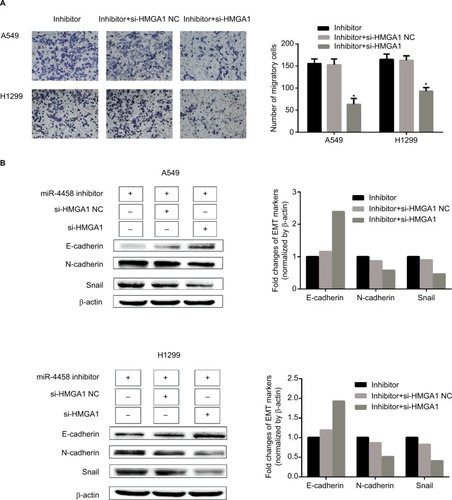
Figure 7 Knockdown of HMGA1 abolished the role of downregulated miR-4458 in the process of migration and EMT.
Notes: (A) A549 and H1299 were cotransfected with miR-4458 inhibitor, si-HMGA1 NC and si-HMGA1, and the transwell results were shown after 48 hours of transfection. (B) Western blot analysis was carried out after 48 hours of transfection.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control.