Figures & data
Table 1 Genomic expression and alterations associated with drug resistance in HNSCC studies
Figure 1 Schematic diagram of signaling pathways that emerging biomarkers TP53, EGFR, PIK3CA, CDKN2A, CCND1, and MET participated in.
Notes: EGFR is a member of the HER family of cell-surface receptor tyrosine kinases. Ligand binding triggers EGFR and activates downstream effectors, thus promoting cell proliferation. Hypoxia induces the expression of EGFR and enhances the phosphorylation of EGFR thereby regulating intrinsic DNA-repair mechanisms, meanwhile EGFR can stabilize HIF-1a. MET is a receptor tyrosine kinase associated with enhanced migration, invasion, and angiogenesis when overexpressed in cancer. PIK3CA encodes p110α, a catalytic subunit of PI3K, which is a heterodimeric kinase with enzymatic activity on lipid and protein substrates. The TP53 has a vital role in the regulation of genes responsible for cell-cycle arrest, senescence, and apoptosis. MDM2 promotes the rapid degradation of the TP53 protein. TP53 missense mutations cause single-amino-acid substitutions that lead to loss of DNA-binding capability. Cyclin D1 is a cell-cycle protein that regulates the key G1-to-S phase transition through formation of complexes with CDKs, such as CDK4 and CDK6. Upon ligand binding, NOTCH receptors undergo a conformational change enabling cleavage and nuclear translocation of the intracellular domain to release the transcriptional repression of downstream target genes.
Abbreviations: CDKs, cyclin-dependent kinases; HIF-1a, hypoxia-inducible factor 1a; PI3K, phosphoinositide 3 kinase.
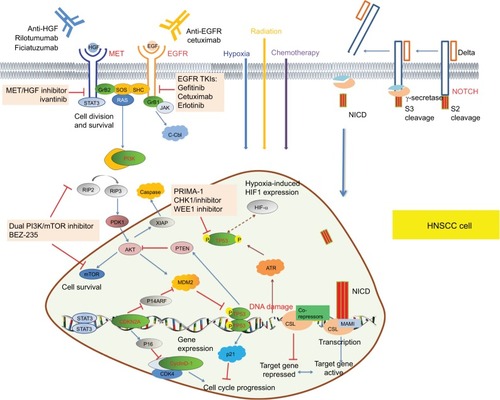
Figure 2 General interactions of genomic mutations (TP53, CDKN2A, CCND1, NOTCH1, PIK3CA, EGFR, and MET) and current target drugs.
Notes: Primary data are from tumor samples with sequencing and CNA data (132 patients/samples) and could be accessed in cBioPortal.Citation6 Detailed description of data mining is (filter neighbors by max 28.8% alteration) found in http://www.cbioportal.org/results/network?Action=Submit&RPPA_SCORE_THRESHOLD=2&Z_SCORE_THRESHOLD=2&cancer_study_list=hnc_mskcc_2016&case_set_id=hnc_mskcc_2016_cnaseq&data_priority=0&gene_list=TP53%2520CDKN2A%2520CCND1%2520NOTCH1%2520PIK3CA%2520EGFR%2520MET&geneset_list=%20&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=hnc_mskcc_2016_gistic&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=hnc_mskcc_2016_mutations&tab_index=tab_visualize.Citation134,Citation135
Abbreviations: US FDA, US Food and Drug Administration.
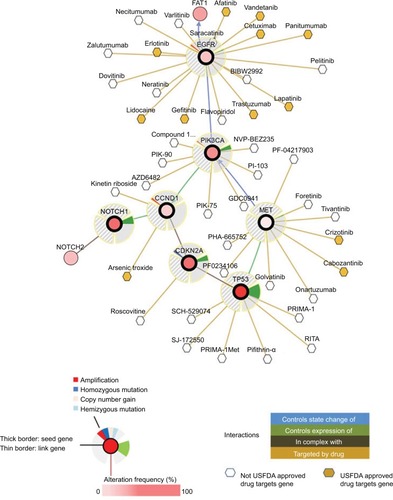
Figure 3 Alteration profiles of the emerging biomarkers from querying 1,627 patients/1,629 samples in 7 studies analyzed in cBioPortal.
Notes: Data from 11,000 cases and all TCGA tumor types from TCGA. Detailed description for data mining could be found in http://www.cbioportal.org/results/cancerTypesSummary?Action=Submit&RPPA_SCORE_THRESHOLD=2&Z_SCORE_THRESHOLD=2&cancer_study_list=hnc_mskcc_2016%2Chnsc_broad%2Chnsc_jhu%2Chnsc_tcga_pub%2Chnsc_tcga_pan_can_atlas_2018%2Chnsc_tcga%2Chnsc_mdanderson_2013&case_set_id=all&data_priority=0&gene_list=TP53%2520CDKN2A%2520CCND1%2520NOTCH1%2520PIK3CA%2520EGFR%2520MET&geneset_list=%20&tab_index=tab_visualize.Citation134,Citation135
Abbreviation: TCGA, The Cancer Genome Atlas.
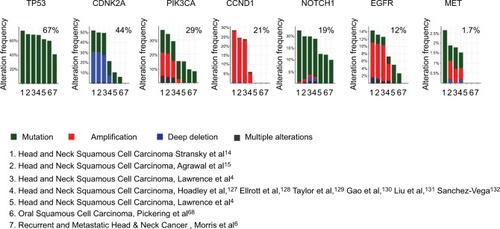
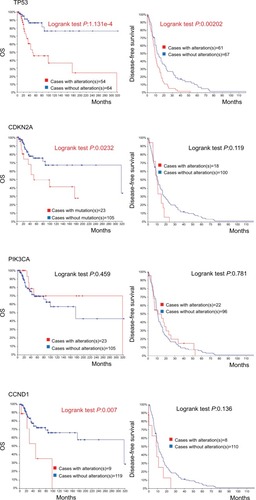
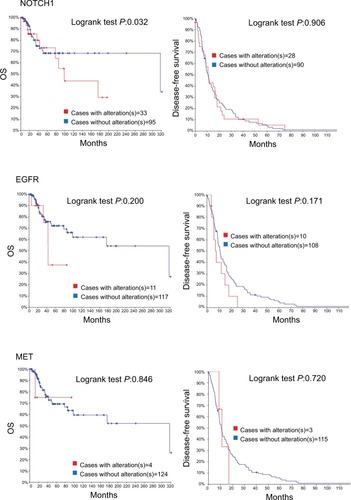
Figure 4 Recurrent and metastatic head and neck cancer samples with sequencing and CNA data (132 patients/samples) analyzed in cBioPortal.
Notes: OS/recurrence-free survival, for cases with/without alteration(s) in query gene. Survival probabilities were calculated with the Kaplan–Meier method, according to the original article.Citation6 Detailed description of data mining could be found in http://www.cbioportal.org/results/survival?Action=Submit&RPPA_SCORE_THRESHOLD=2&Z_SCORE_THRESHOLD=2&cancer_study_list=hnc_mskcc_2016&case_set_id=hnc_mskcc_2016_cnaseq&data_priority=0&gene_list=TP53&geneset_list=%20&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=hnc_mskcc_2016_gistic&genetic_profile_ids_PROFILE_MUTATION_EXTENDED=hnc_mskcc_2016_mutations&tab_index=tab_visualize.Citation134,Citation135
Abbreviation: OS, overall survival.