Figures & data
Figure 1 Volcano plot for lncRNAs between early and late stages of colorectal cancer samples.
Notes: Each gene is marked as a dot; those genes that were significantly upregulated are highlighted in red, the downregulated genes are highlighted in blue, and the nonsignificant genes are labeled as gray dots. In the figure, the gray lines indicate the marginal lines separating differently expressed lncRNAs from nondifferently expressed lncRNA, with the horizontal lines denoting the P-value threshold (FDR <0.01) and the vertical lines denoting the FC cutoff (|logFC|≥ 1.5).
Abbreviations: DEG, differentially expressed gene; FC, fold change; FDR, false discovery rate; lncRNAs, long noncoding RNAs.
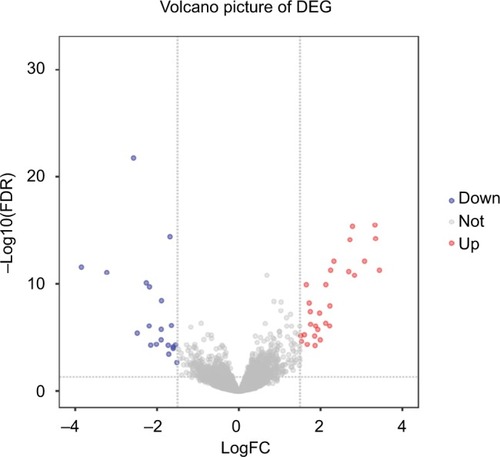
Figure 2 Heat map of differentially expressed lncRNAs in colorectal cancer samples.
Notes: Each block represents the color-coded expression levels of differentially expressed lncRNAs in colorectal cancer samples. Orange represents the relatively higher expression, whereas the blue represents the relatively lower expression.
Abbreviation: lncRNAs, long noncoding RNAs.

Figure 3 Survival analysis of ten lncRNAs.
Notes: The red line of each image represents the samples with the high expression (expression higher than median) of the corresponding lncRNA, whereas blue line represents the samples with the low expression (expression lower than median) of the corresponding lncRNA. Log-rank test was utilized to perform the statistical analysis.
Abbreviation: lncRNAs, long noncoding RNAs.
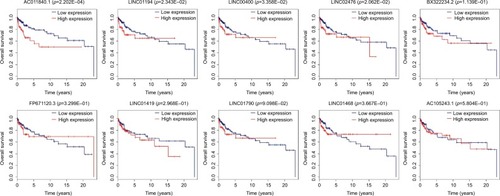
Figure 4 Expression of Linc01194 in colorectal cancer cell lines by real-time-PCR.
Notes: (A) Expression of Linc01194 in colorectal cancer cell lines. (B) Interference efficiency of si-Linc01194 in HCT116 cells. ***P<0.001.
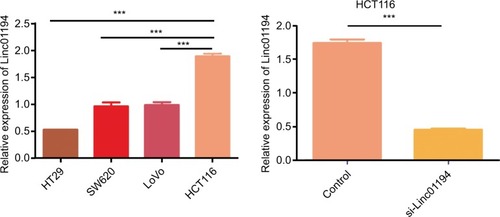
Figure 5 Impact of Linc01194 on HCT116 cells.
Notes: (A) Proliferation activity of HCT116 cells measured by CCK-8 assay. After transfecting with negative control or si-Linc01194, the absorbance at 450 nm was detected at 24, 48, 72, and 96 hours. (B) Migration and invasion of HCT116 cells. After transfecting with negative control or si-Linc01194, cells that crossed the membrane were stained with crystal violet and observed under a microscope (200×). (C) Quantification of the invasion and migration assays. Significant reduction was observed in si-Linc01194 group compared to negative control group migration and invasion assays (Student’s t-test). (D) HCT116 transfected with si-Linc01194 cells were significantly arrested in G0/G1 phase in cell cycle. **P<0.01; ***P<0.001.
Abbreviation: CCK-8, cell counting-kit-8.
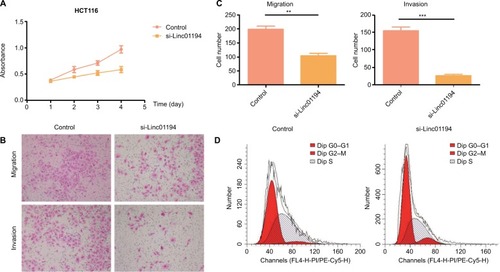
Figure 6 Western blot of N-cadherin, vimentin, E-cadherin, and β-actin.
Notes: After transfecting with negative control or si-Linc01194, protein of HCT116 cells were extracted for Western blot analysis.
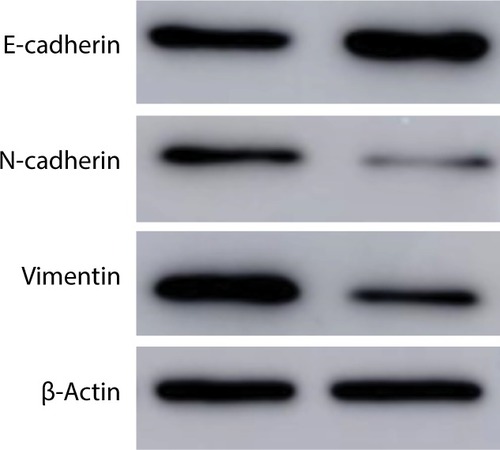
Table 1 Clinical parameters of colorectal cancer patients
Table 2 Relationship between Linc01194 expression and clinical parameters
Table 3 Gene Ontology analysis of mRNA involved in the ceRNA network of Linc01194
Figure 7 The role Linc01194 plays in colorectal cancer.
Notes: Linc01194 could promote proliferation, invasion, and EMT in colon cancer.
Abbreviation: EMT, epithelial–mesenchymal transition.
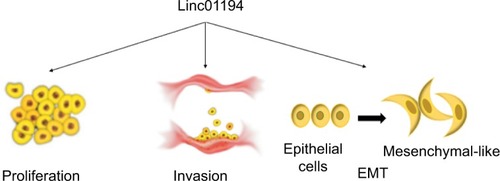
Table S1 Fifty-one differently expressed lncRNAs in colon cancer
Table S2 mRNA involved in the ceRNA network of Linc01194
