Figures & data
Figure 1 JAM3 was hypermethylated in primary CRC tissues, plasma, and cell lines.
Notes: (A) Expression level of JAM3 was significantly downregulated in CRC tissues compared to that in normal controls in the Skrzypczak colorectal dataset and TCGA expression array dataset from Oncomine. (B) A significant difference in JAM3 methylation status was observed between 125 CRC tissues and 29 adjacent normal tissues in the GSE25062 dataset. (C) Schematic structure of the JAM3 promoter CpG islands, CpG sites (short vertical lines), and qMSP region is indicated (from forward primer to reverse primer). The transcription start site is shown by a curved arrow. (D) Methylation percentage of JAM3 in 24 paired CRC tissues and normal tissues was detected by qMSP. (E) Methylation percentage of JAM3 in plasma from 18 CRC cases and 18 healthy subjects was examined by qMSP (P<0.001, two-sided Student’s t-test). (F) Methylation percentage of JAM3 in CRC cell lines (HT29, HCT116, SW480, and SW620) and normal colonic NCM460 cells was detected by qMSP (P<0.001, two-sided Mann–Whitney rank-sum test). (G) ROC curves showed the performance of JAM3 methylation status from CRC tissues in predicting CRC. (H) ROC curves showed the performance of JAM3 methylation status from CRC plasma in predicting CRC. Sensitivity means true-positive rate. Specificity means false-positive rate. Results are shown as mean ± SD. ***P<0.001.
Abbreviations: CRC, colorectal cancer; TCGA, The Cancer Genome Altas; qMSP, quantitative methylation-specific PCR; ROC, receiver-operating characteristic.
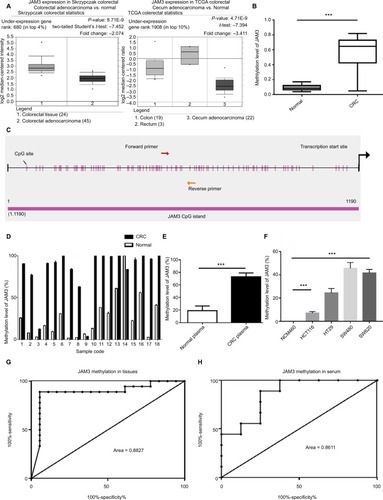
Figure 2 JAM3 was downregulated in CRC tissues, plasma, and cell lines.
Notes: (A) JAM3 mRNA expression levels in 24 paired CRC tissues and paired normal tissues were detected using qPCR, relative to the value of GAPDH in each sample (P<0.05, two-sided Student’s t-test). (B) ROC curves showed the performance of JAM3 expression levels from tissues in predicting CRC. (C) JAM3 mRNA expression levels in 18 CRC and as well as paired normal plasma samples were detected using qPCR (P<0.05, two-sided Student’s t-test). (D) ROC curves showed the performance of JAM3 expression levels from CRC tissues in predicting CRC. (E) Representative Western blot image for protein expression level of JAM3 in CRC tissues and paired normal tissues. (F) Western blot image for protein expression level of JAM3 in CRC cell lines (HT29, HCT116, SW480, and SW620) and normal colonic NCM460 cells. (G) JAM3 mRNA expression levels in CRC cell lines (HT29, HCT116, SW480, and SW620) and normal colorectal epithelial NCM460 cells were detected using qPCR (P<0.001, two-sided Mann–Whitney rank-sum test). (H) Representative images of JAM3 staining in CRC tissues and paired normal tissues. Normal tissues showed strong positive staining, whereas CRC tissue displayed weak staining. The right panel (magnification ×400) was magnification of where inset in the left tissue array (magnification ×100). Results are shown as mean ± SD. *P<0.05 and ***P<0.001.
Abbreviations: CRC, colorectal cancer; qPCR, quantitative PCR; ROC, receiver-operating characteristic.
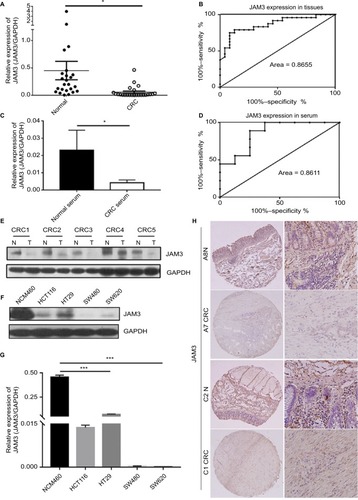
Figure 3 Demethylating agent 5-AZA treatment induced JAM3 expression in CRC cell lines.
Notes: (A) JAM3 methylation status after 5 µm 5-AZA treatment in CRC cell lines (HT29, HCT116, SW480, and SW620) and normal colorectal NCM460 cells are measured by qMSP. (B) qPCR and (C) Western blots were used to examine mRNA and protein expression levels of JAM3 after 5 µm 5-AZA treatment in CRC cell lines and NCM460 cells. Results are shown as mean ± SD (two-sided Student’s t-test). *P<0.05 and **P<0.01.
Abbreviations: 5-AZA, 5-Aza-2′-deoxycytidine; CRC, colorectal cancer; qMSP, quantitative methylation-specific PCR; qPCR, quantitative PCR.
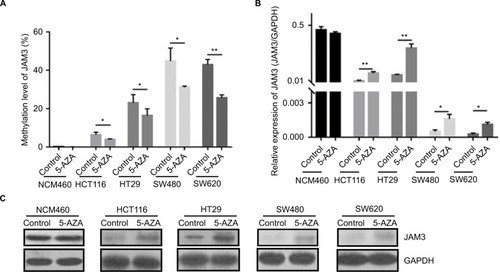
Table 1 Association between clinicopathologic characteristics and JAM3 methylation status
Figure 4 Restoration of JAM3 suppressed CRC cell growth and migration.
Notes: (A) Expression of JAM3 in SW620 and HT29 cells transfected with JAM3 expression vector, and control vector was confirmed by Western blot. (B) JAM3 expression significantly suppressed the number of viable cells of JAM3-transfected SW620 and HT29 cells compared to that of control vector-transfected cells (P<0.01, two-sided Student’s t-test). (C) Representative result of transwell assay in JAM3-transfecting as well as control vector transfecting HT29 and SW620 cells. (D) Representative result of monolayer scratch-healing assay in JAM3-transfecting as well as control vector transfecting SW620 and HT29 cells. (E) Representative result of cell-cycle analysis in JAM3-transfecting as well as control vector transfecting SW620 and HT29 cells. Results are shown as mean ± SD. **P<0.01.
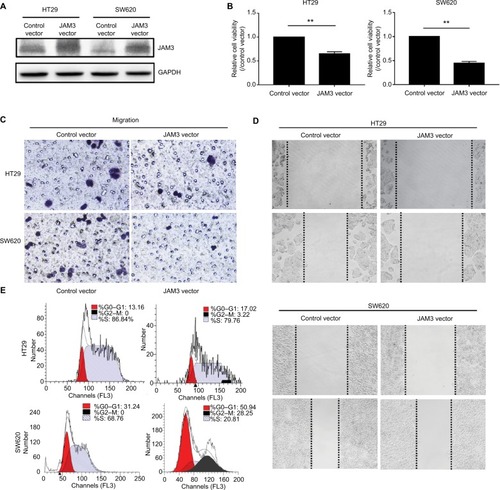
Figure 5 JAM3 suppression promoted cell migration and reduced apoptosis in colonic NCM460 cells.
Notes: JAM3 mRNA (A) and protein (B) expressions were efficiently inhibited in NCM460 cells transfected with siJAM3. GAPDH was used as an internal control (P<0.01, two-sided Student’s t-test). (C) Representative result of CCK8 assay (P<0.01, two-sided Student’s t-test). (D) Representative colony formation assays in NCM460 cells transfected with siJAM3 and siNC. (E) Representative transwell assays in NCM460 cells transfected with siJAM3 and siNC. (F) Cell-cycle distribution of NCM460 cells transfected with siJAM3 and siNC was detected by flow cytometry analysis. (G) Representative result of cell apoptosis analysis. **P<0.01 and ***P<0.001.
Abbreviations: CCK8, Cell Counting Kit-8; siJAM3, siRNA–JAM3; siNC, siRNA control; FITC, fluorescein isothiocyanate.
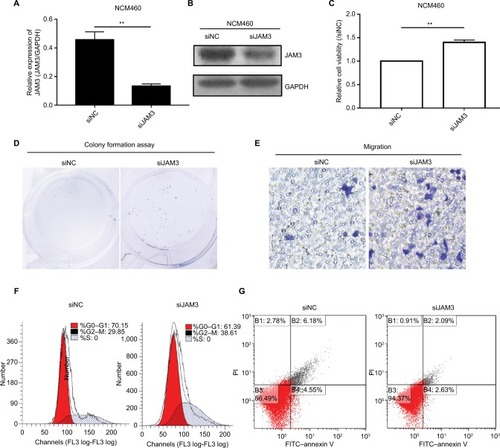
Figure 6 JAM3 participated in EMT in colonic epithelial cells.
Notes: Molecular interaction networks involving JAM3 (A) and TJP1 (B) were generated by using GCBI database and GeneCards database. (C) Protein expression levels of Bcl2, caspase-3, P21, P27, cyclin D1, E-cadherin, vimentin, TJP1, p-β-catenin, and β-catenin in NCM460 cells transfected with siJAM3 and siNC. (D) Protein expression levels of Bcl2, caspase-3, P21, P27, cyclin D1, E-cadherin, vimentin, TJP1, p-β-catenin, and β-catenin in JAM3-transfecting as well as control vector transfecting SW620 and HT29 cells.
Abbreviations: EMT, epithelial–mesenchymal transition; siNC, siRNA control; GCBI, Gene-Cloud of Biotechnology Information.
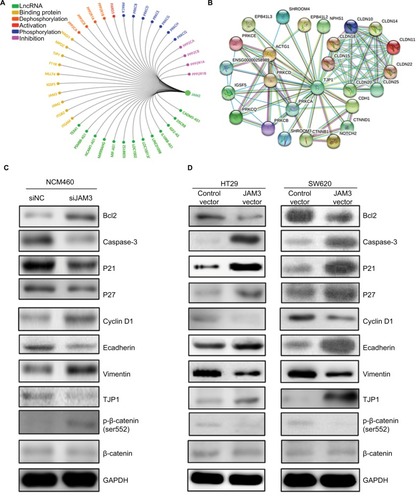
Table S1 Clinical characteristics of patients with CRC and normal controls
Table S2 Clinical characteristics of patients with CRC and normal controls in tissue array
