Figures & data
Figure 1 The flowchart of our study design.
Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA; RT-qPCR, quantitative real-time PCR.
Figure 2 ROC curve analysis for the detection of CRC using piR-001311 (A), piR-004153 (B), piR-017723 (C), piR-017724 (D), piR-020365 (E) and the piRNA-based Panel I (F) in the training set.
Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA; ROC, receiver operating characteristic.
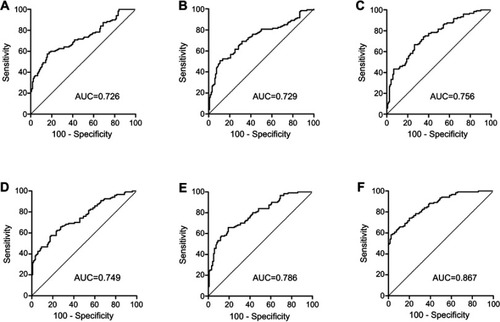
Table 1 The selected serum piRNA expression levels in patients with CRC and healthy controls in training set and validation set (median [IQR])
Figure 3 Differential presence of serum piRNAs in the validation set. The expression levels of piR-001311 (A), piR-004153 (B), piR-017723 (C), piR-017724 (D), piR-020365 (E) and piRNA-based Panel I (F) were determined by RT-qPCR in 100 CRC patients, 20 CRA patients and 100 healthy controls. * P<0.05.
Abbreviations: CRA, colorectal adenoma; CRC, colorectal cancer; piRNA, Piwi-interacting RNA; RT-qPCR, quantitative real-time PCR.
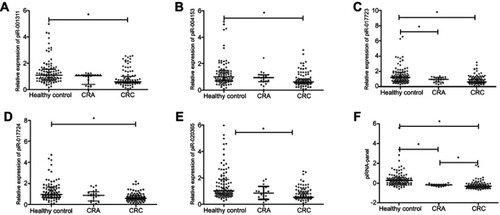
Table 2 Performance of piRNA-based panel I and CEA-based panel II in the detection of CRC from healthy controls
Figure 4 ROC for piRNA-based Panel I and CEA-based Panel II in the training set (A), validation set (B) and combination set (C).
Abbreviations: AUC, area under the ROC curve; CEA, carcinoembryonic antigen; piRNA, Piwi-interactingRNA; ROC, receiver operating characteristic.
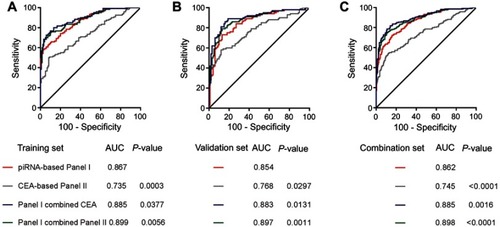
Figure 5 Fagan’s nomogram for the calculation of the probability that an individual has CRC based on the piRNA-based Panel I.
Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA.
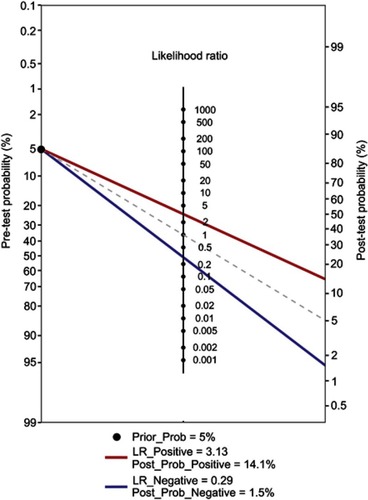
Figure 6 Kaplan–Meier curves for OS (A) and PFS (B) according to piR-017724 level in CRC patients. The median piR-017724 expression level in the tumor samples was chosen as the cutoff point.
Abbreviations: CRC, colorectal cancer; OS, overall survival; PFS, progression-free survival.
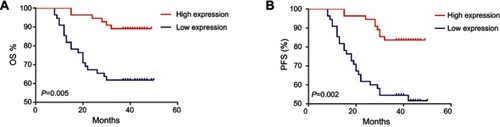
Table 3 Cox proportional hazards regression model analysis for OS and PFS in CRC patients
Figure S1 The expressions of reference genes and piRNAs between CRC patients and healthy controls in the training set. (A) Ct values of miR-191-5p, U6 and combination of miR-191-5p and U6 in CRC patients and healthy controls (all *P>0.05). No significant difference was found within three reference genes in both groups. (B–F) The expression levels of piR-001311 (B), piR-004153 (C), piR-017723 (D), piR-017724 (E) and piR-020365 (F) were determined by RT-qPCR in 120 CRC patients and 120 healthy control.
Abbreviations: CRC, colorectal cancer; piRNA, Piwi-interacting RNA.
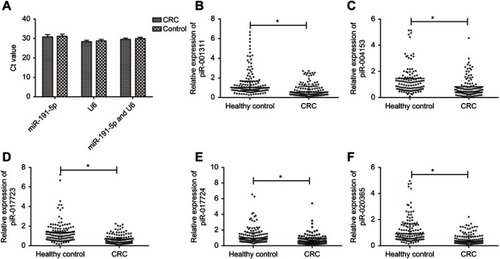
Figure S2 Stability of serum piRNA expressions. Serum levels of piR-001311 (A), piR-004153 (B), piR-017723 (C), piR-017724 (D) and piR-020365 (E) remained stable when treated with prolonged incubation at room-temperature, repetitive freeze-thaw cycles, or prolonged incubation at –80℃.
Abbreviation: piRNA, Piwi-interacting RNA.
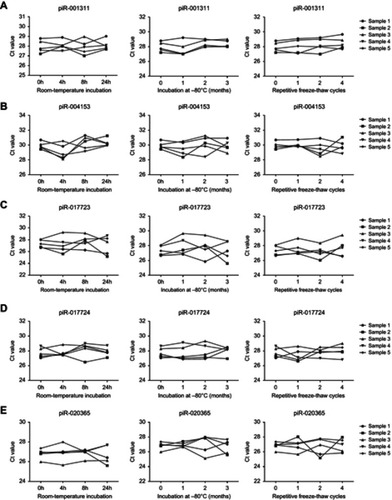
Figure S3 ROC curve analysis for the detection of CRC using piR-001311 (A), piR-004153 (B), piR-017723 (C), piR-017724 (D), piR-020365 (E) and the piRNA-based panel (F) in the validation set.Abbreviations: AUC, area under the ROC curve; CRC, colorectal cancer; piRNA, Piwi-interacting RNA; ROC, receiver operating characteristic.
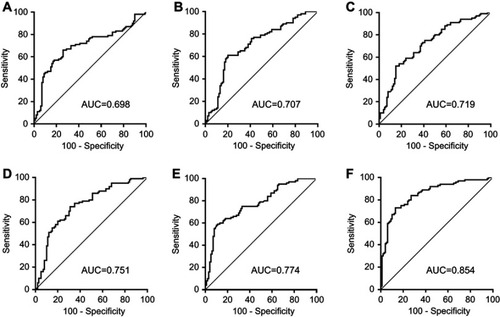
Figure S4 ROC curve analysis for the diagnostic performance of CEA (A), CA19-9 (B) and CEA-based panel II (C) in the training set.
Abbreviations: AUC, area under the ROC curve; CEA, carcinoembryonic antigen; ROC, receiver operating characteristic.
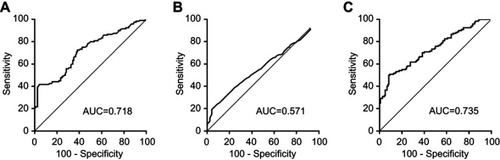
Table S1 Characteristics of participants enrolled in the study
Table S2 Selected piRNAs that were significantly downregulated in serum samples of CRC patients compared with healthy controls.The values showing in the columns referred to RPM value
Table S3 Diagnostic performance of individual biomarkers in the training, validation and combination sets
