Figures & data
Figure 1 LAPTM4B enhances HCC growth in vitro and in vivo. (A) Construction of stably LAPTM4B knockdown SMMC-7721 cells (shLAPTM4B). The knockdown efficacy was determined by IF (left) and qRT-PCR (right). Representative images are shown. (B) The colony formation of control and LAPTM4B knockdown SMMC-7721 cells. Representative images are shown. (C) The proliferation of control and LAPTM4B knockdown SMMC-7721 cells were detected by CCK-8 assay. (D) Construction of stably LAPTM4B overexpressing PLC/PRF/5 cells. The overexpression efficacy was determined by IF (left) and qRT-PCR (right). Representative images are shown. (E) The colony formation of control and LAPTM4B overexpressing PLC/PRF/5 cells. Representative images are shown. (F) The proliferation of control and LAPTM4B overexpressing PLC/PRF/5 cells was detected by CCK-8 assay. (G) SMMC-7721 cells stably expressing LAPTM4B shRNA or the negative control were used for in vivo tumorigenesis. (H) PLC/PRF/5 cells stably overexpressing LAPTM4B or the negative control were used for in vivo tumorigenesis. Data are shown as mean ± SD, *P<0.05, **P<0.01.
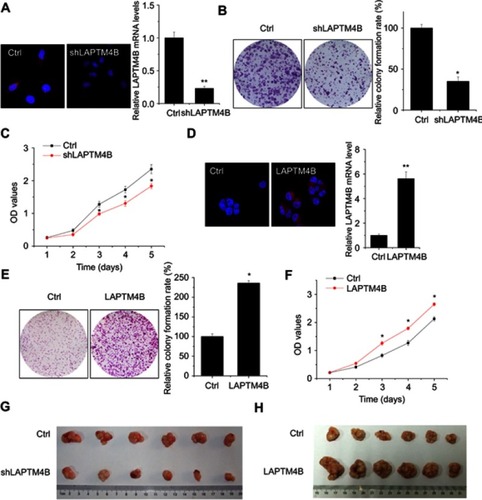
Figure 2 LAPTM4B enhances survival and inhibits apoptosis under starvation conditions. (A) Control and LAPTM4B knockdown SMMC-7721 cells were treated with HBSS. At different time point, the cell survival was detected by CCK-8 assay. (B) Control and LAPTM4B overexpressing PLC/PRF/5 cells were treated with HBSS. At different time point, the cell survival was detected by CCK-8 assay. (C) Control and LAPTM4B knockdown SMMC-7721 cells were treated with HBSS for 24 hrs. The apoptosis was detected by flow cytometry. (D) Control and LAPTM4B overexpressing PLC/PRF/5 cells were treated with HBSS for 24 hrs. The apoptosis was detected by flow cytometry. Data are shown as mean ± SD, *P<0.05.
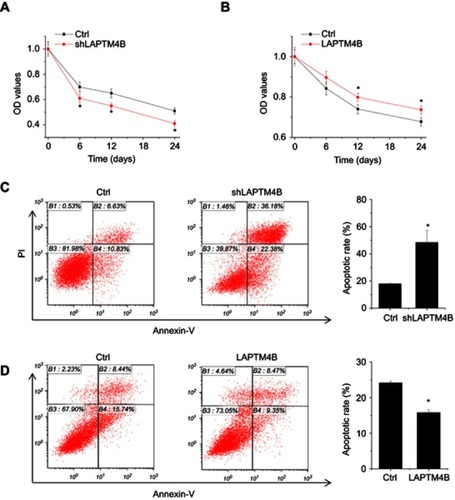
Figure 3 LAPTM4B induces autophagy in HCC cells. (A) Evaluation for LC3-Ⅱand p62 alteration indicative of autophagy induction in control and LAPTM4B knockdown SMMC-7721 cells treated with EBSS for 24 hrs (left). Densitometric analysis normalized to ACTB demonstrating the effect of LAPTM4B silencing on LC3-II levels (right). (B) Evaluation for LC3-Ⅱand p62 alteration indicative of autophagy induction in control and LAPTM4B overexpressing PLC/PRF/5 cells treated with EBSS for 24 hrs (left). Densitometric analysis normalized to ACTB demonstrating the effect of LAPTM4B overexpression on LC3-II levels(right). (C) Representative immunofluorescent images showing redistribution of autophagic marker LC3 in LAPTM4B knockdown SMMC-7721 cells treated with EBSS for 24 hrs were taken on a confocal microscope (left). The average number of LC3 dots per cell was counted in more than 5 fields with at least 100 cells for each group (right). Representative images are shown. (D) Representative immunofluorescent images showing redistribution of autophagic marker LC3 in LAPTM4B overexpressing PLC/PRF/5 cells treated with EBSS for 24 hrs were taken on a confocal microscope (left). The average number of LC3 dots per cell was counted in more than 5 fields with at least 100 cells for each group (right). Representative images are shown. Data are shown as mean ± SD, *P<0.05.
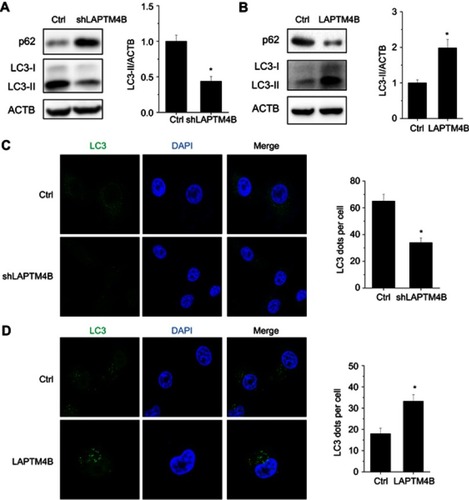
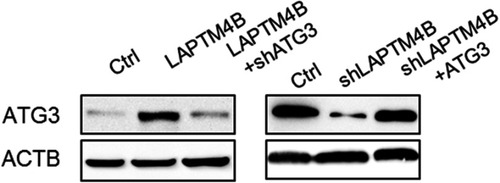
Figure 4 LAPTM4B upregulates ATG3 expression. (A) Hierarchically clustered heatmap of upregulated and downregulated genes in control and LAPTM4B knockdown SMMC-7721 cells treated with HBSS for 24 hrs. (B) The top 10 biological processes affected by LAPTM4B downregulation by GO. (C) The mRNA and protein levels of ATG3 in control and LAPTM4B knockdown SMMC-7721 cells. (D) The mRNA and protein levels of ATG3 in control and LAPTM4B overexpressing PLC/PRF/5 cells. (E) ATG3 protein levels in the xenografts of nude mice injected with control and LAPTM4B knockdown SMMC-7721 cells were analyzed by performing western blotting (left). Quantification of ATG3 protein levels (right). (F) ATG3 protein levels in the xenografts of nude mice injected with control and LAPTM4B overexpressing PLC/PRF/5 cells was analyzed by performing western blotting (left). Quantification of ATG3 protein levels (right). Data are shown as mean ± SD, *P<0.05.
Figure 5 ATG3 takes part in LAPTM4B-mediated autophagy and survival. (A, B) The effect of ATG3 overexpression on LC3 accumulation (A) and apoptosis (B) mediated by LAPTM4B knockdown. (C, D) The effect of ATG3 knockdown on LC3 accumulation (C) and apoptosis (D) mediated by LAPTM4B overexpression. Data are shown as mean ± SD, *P<0.05.
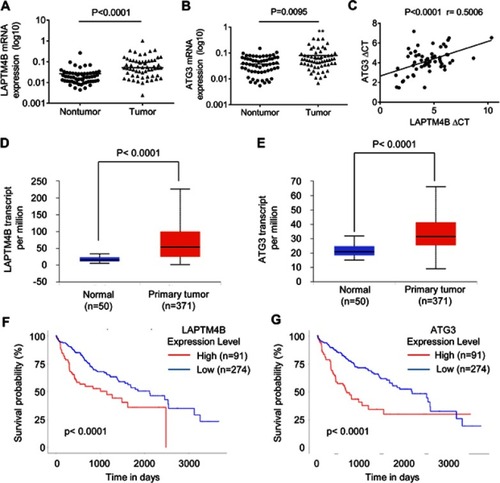
Figure 6 A positive correlation between LAPTM4B and ATG3 expression in HCC tissues. (A) The LAPTM4B mRNA levels in 76 pairs of HCC and corresponding adjacent noncancerous tissues were analyzed by qRT-PCR. (B) The ATG3 mRNA levels in 76 pairs of HCC and corresponding adjacent noncancerous tissues were analyzed by qRT-PCR. (C) The correlation between LAPTM4B and ATG3 mRNA expression in 76 HCC tissues. (D) LAPTM4B mRNA expression in normal and HCC tissues from TCGA datasets. Data are mean ± SD. (E) ATG3 mRNA expression in normal and HCC tissues from TCGA datasets. Data are mean ± SD. (F) Kaplan-Meier survival analysis of the whole HCC patients according to LAPTM4B mRNA expression. The 3rd quatile of LAPTM4B expression in patients was used as a cutoff. High expression patients show expression value >3rd quatile, and low expression patients show expression value <3rd quatile. (G) Kaplan-Meier survival analysis of the whole HCC patients according to ATG3 mRNA expression. The 3rd quatile of ATG3 expression in patients was used as a cutoff.