Figures & data
Figure 1 The negative regulators of JAK-STAT signaling. Binding of the ligand to cytokine receptor induces receptor dimerization and activation of receptor associated JAK kinase, which in turn phosphorylates STAT proteins. After forming a homodimer, STAT proteins translocate to the nucleus to control gene expression. Negative regulation of the JAK-STAT pathway is provided by PTPs, SOCS and PIAS proteins.
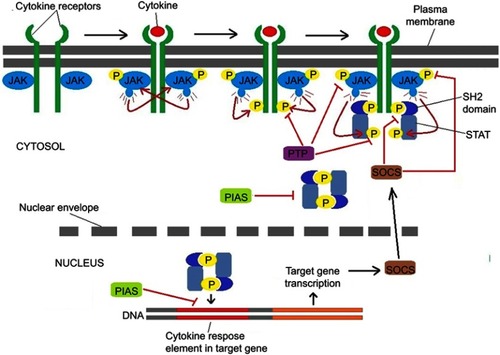
Figure 2 Structure characteristics of STATs, PIAS and SOCS. (A) Structure of STAT. ND, N domain; CCD, coiled coil domain; DBD, DNA-binding domain; LD, linker domain; SH2, Src homology 2 domain; TAD, transactivation domain. (B) The domain structure of PIAS proteins. SAP/LXXLL, SAF (scaffold attachment factor A and B) -A/B, Acinus and PIAS domain (it can recognize and bind to AT-rich DNA sequences), Within the SAP domain, there is a conserved LXXLL (it mediates interactions between nuclear receptors and their co-regulators) signature motif; PINIT, Pro-Ile-Asn-Ile-Thr motif (a highly conserved region of PIAS proteins and it may be involved in the nuclear retention of PIAS3); RLD, RING finger-like zinc binding domain; AD/SIM, acidic domain (AD), Within the AD, there is a putative SUMO1 interaction motif (SIM) in all PIASs except PIASy; S/T, serine-threonine rich region (in all PIASs except PIASy). (C) Structure of SOCS proteins. All eight members are characterized by their N-terminal region with variable length and limited homology, an extended SH2 domain (ESS), a central SH2 domain and a conserved SOCS box at the C-terminus.
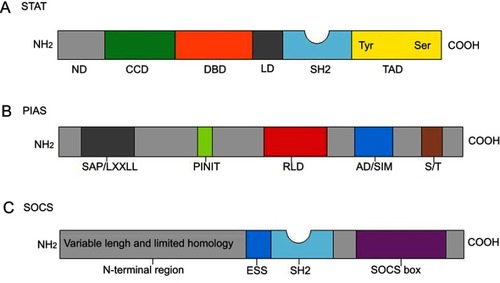
Table 1 Direct inhibitors of STAT3
