Figures & data
Figure 1 miR-193a-3p was found downregulated in CRC cells compared with normal colonic epithelial cell through qRT-PCR analysis. Data were shown as mean ± SD. **P<0.01.
Abbreviations: CRC,colorectal cancer; qRT-PCR, quantitative real-time PCR.
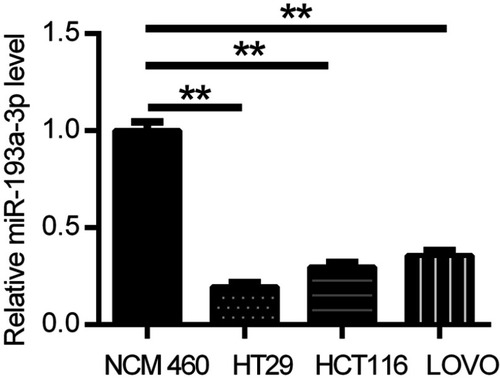
Figure 2 miR-193a-3p suppressed HT-29 cell proliferation, migration and angiogenesis in vitro. (A) The expression of miR-193a-3p was inclined or decreased in HT-29 cell via transfecting with miR-193a-3p mimic or inhibitor. (B) CCK-8 assay was performed to detect the viability of HT-29 cell after transfecting with miR-193a-3p mimic or inhibitor. (C) Transwell assay was performed to assess the migration ability of HT-29 cell after transfecting with miR-193a-3p mimic or inhibitor. (D) HUVEC were cultured on Matrigel- counted plate with CM from HT-29 cell transfecting with miR-193a-3p mimic or inhibitor. Data were shown as mean ± SD. **P<0.01,*** P<0.001.
Abbreviations: CRC,colorectal cancer; CCK-8, cell counting kit-8; CM, conditional medium; HUVEC, human umbilical vein endothelial cell.
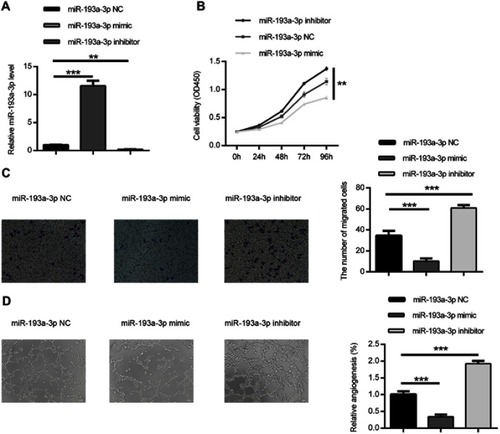
Figure 3 PLAU was confirmed as a direct target gene of miR-193a-3p. (A) PLAU was predicted as a target gene of miR-193a-3p according to the bioinformatic method. (B) HT-29 cell was contransfected with miR-193a-3p-NC or miR-193a-3p-mimic and WT or Mut-type PLAU-3ʹUTR reporter plasmid. Luciferase activity was measured 48 hrs after transfection. (C) PLAU protein expression levels were measured in HT-29 cell after transfection by Western blot analysis. Data were shown as mean ± SD. **P<0.01, ***P<0.001.
Abbreviations: NC, negative control; UTR, untranslated region; WT, wild type; Mut, mutant.
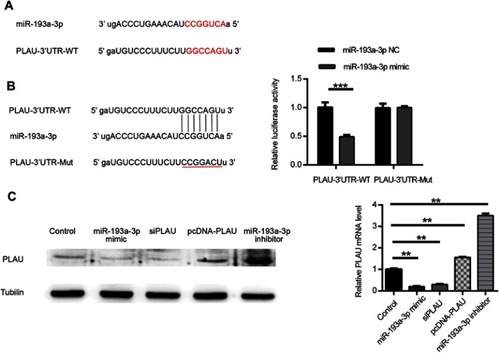
Figure 4 Biological effect of PLAU on the proliferation, migration and angiogenesis ability in HT-29 cell line. (A) The expression of PLAU in CRC tissues and normal colorectal tissues in TCGA datasets. (B) CCK-8 assay was performed to detect the viability of HT-29 cell after transfecting with PLAU plasmid or inhibitor. (C) Transwell assay performed to assess the invasion ability of HT-29 cell after transfecting with PLAU plasmid or inhibitor. (D) HUVEC were cultured on Matrigel-counted plate with CM from HT-29 cell transfecting with PLAU plasmid or inhibitor. Data were shown as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: COAD, colon adenocarcinoma; HUVEC, human umbilical vein endothelial cell; CM, conditional medium.
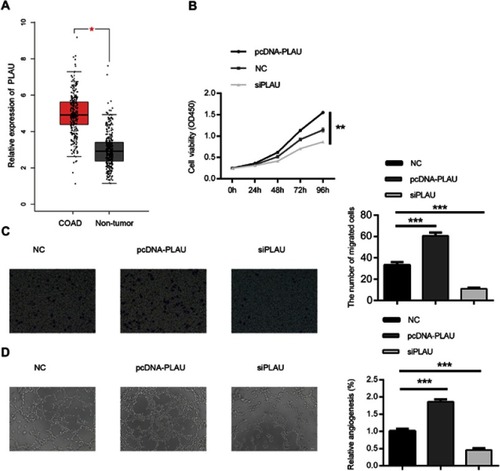
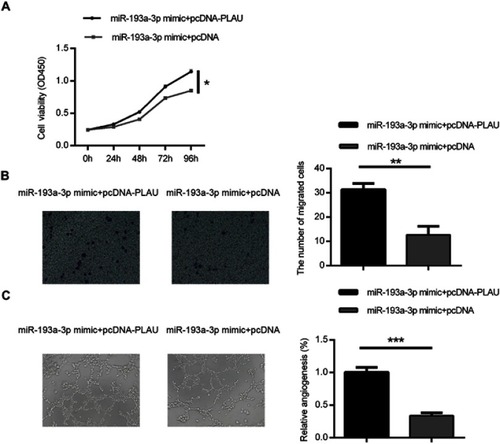
Figure 5 Forced PLAU over-expression could rescue the inhibitory effect of miR-193a-3p on CRC cells. Cell proliferation (A), migration (B) and angiogenesis (C) were measured in HT-29 cells co-transfected with miR-193a-3p mimic and PLAU plasmid or blank vector. Data were shown as mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviation: CRC, colorectal cancer.