Figures & data
Figure 1 Expression level of hsa_circRNA_102958 was upregulated in CRC. (A) Relative expression of hsa_circRNA_102958 in 58 paired colorectal cancer (CRC) tissues and normal controls by quantitative real-time PCR. (B) In situ hybridization (ISH) was used to analyze hsa_circRNA_102958 expression in paired CRC tissues and normal controls. Scale bar, 50 μm. (C) Expression of hsa_circRNA_102958 in Stages I/II and Stages III/IV was determined. (D) Expression of hsa_circRNA_102958 in CRC tissues with lymph node metastasis (N1/N2) or not (N0) was analyzed. (E) Differential expression patterns of hsa_circRNA_102958 in CRC cell lines and FHC cells by quantitative real-time PCR. (F) High hsa_circRNA_102958 expression indicated low survival rate in CRC patients. *P<0.05, **P<0.01 and ***P<0.001.
Figure 2 Loss of hsa_circRNA_102958 suppressed proliferation, migration and invasion of colorectal cancer (CRC) cells. (A) Hsa_circRNA_102958 expression was decreased in SW480 and HCT116 cells after transfection with si-circRNA. (B and C) The proliferation was inhibited after hsa_circRNA_102958 silencing as shown by CCK8 and colony formation assays. (D and E) Transwell assay indicated that hsa_circRNA_102958 knockdown suppressed the migration and invasion of SW480 and HCT116 cells. (F) Xenograft experiment indicated that tumor volumes were reduced after hsa_circRNA_102958 silencing. *P<0.05 and **P<0.01.
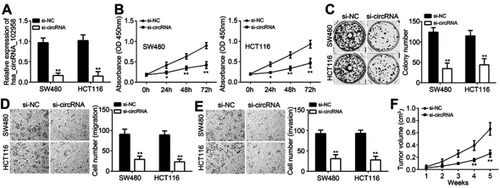
Figure 3 Hsa_circRNA_102958 was a sponge for miR-585 and miR-585 targeted CDC25B. (A) Analysis of binding sites with miR-585 in hsa_circRNA_102958. (B) Luciferase reporter assay indicated that hsa_circRNA_102958-WT reporter activity was inhibited by miR-585 mimics. (C) RNA pulldown assay indicated that biotin-labeled miR-585-WT precipitated hsa_circRNA_102958 in SW480 and HCT116 cells. (D) Hsa_circRNA_102958 silencing promoted miR-585 expression in SW480 and HCT116 cells. (E) miR-585 expression was downregulated in colorectal cancer (CRC) tissues. (F) miR-585 expression was negatively correlated with hsa_circRNA_102958 in CRC tissues. (G) Analysis of binding sites with miR-585 in CDC25B 3ʹ-UTR. (H) Luciferase reporter assay indicated that CDC25B 3ʹ-UTR-WT reporter activity was inhibited by miR-585 mimics. (I and J) CDC25B expression was suppressed by miR-585 mimics. (K) CDC25B expression was upregulated in CRC tissues by quantitative real-time PCR. (L) Based on TCGA data, CDC25B expression was increased in CRC tissues. (M) CDC25B expression was reversely correlated with miR-585 in CRC tissues. **P<0.01 and ***P<0.001.
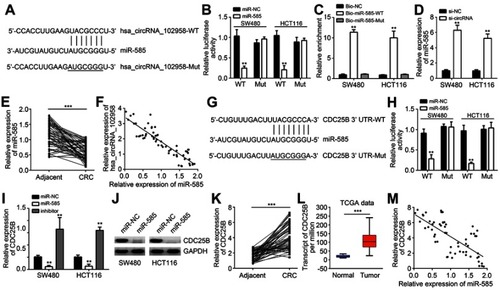
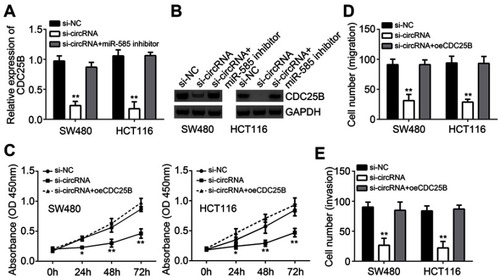
Figure 4 Hsa_circRNA_102958 promoted CRC progression through CDC25B. (A and B) CDC25B expression was detected in SW480 and HCT116 cells transfected with siRNA or miR-585 inhibitors. (C–E) CCK8 assay showed that CDC25B overexpression reversed the effects of hsa_circRNA_102958 silencing on proliferation, migration and invasion. **P<0.01.