Figures & data
Table 1 Clinical Parameters of Patients with Breast Cancer in This Study
Table 2 The Sequences of the Primers in This Study
Figure 1 CXCL11 and miR-1298-5p were identified as our interested mRNA and miRNA. (A) The top 10 most upregulated DEGs from GSE139038 data analysis are shown. GSE139038 is the gene microarray profile of BC. Limma was performed to screen out the DEGs with an adj. (adjusted) P value smaller than 0.05 and a logFC (fold change) bigger than 0. DEGs: differentially expressed genes. (B) The expression of CXCL11 in 1085 tumor and 291 normal breast samples was assessed by GEPIA. BRCA, breast invasive carcinoma. *P<0.01. (C) The effect of CXCL11 on survival of BC patients was analyzed by Kaplan Meier Plotter (http://kmplot.com). (D) The overlapping miRNAs between the dataset of the miRNAs that target CXCL11 predicted by starBase (http://starbase.sysu.edu.cn/) and the dataset of downregulated DEGs from GSE143564 analysis. GSE143564 is the miRNA microarray profile of BC. Limma was applied to analyze the downregulated miRNAs in BC with P < 0.05 and logFC < 0.
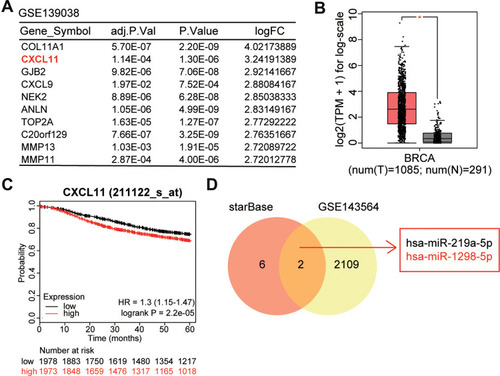
Figure 2 The expression of miR-1298-5p and CXCL11 was detected in tissues and cell lines. (A) The expression of miR-1298-5p in BC tissues and the normal breast tissues was measured by RT-qPCR. (B) The expression of CXCL11 mRNA in BC tissues and the normal breast tissues was measured by RT-qPCR. (C) The relationship between miR-1298-5p expression and CXCL11 mRNA expression was analysed. (D) The expression of miR-1298-5p in BC cell lines (T-47D, SK-BR-3, MDA-MB-231, MCF7, BT-474) and normal cell line (MCF10A) was measured by RT-qPCR. *P<0.05, **P<0.001 versus MCF10A. (E) The expression of CXCL11 mRNA in BC cell lines (T-47D, SK-BR-3, MDA-MB-231, MCF7, BT-474) and normal cell line (MCF10A) was measured by RT-qPCR. *P<0.05, **P<0.001 versus MCF10A.
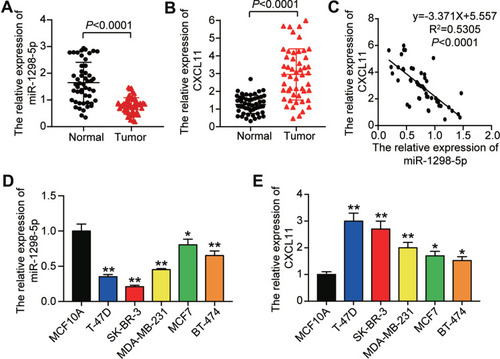
Figure 3 miR-1298-5p inhibited the cell viability, proliferation, migration, and adhesion in BC cells. (A) The expression of miR-1298-5p in T-47D and SK-BR-3 cells transfected with miR-1298-5p mimic or negative control was detected by RT-qPCR. (B) The cell viability was measured by CCK-8 assay after the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic or negative control on Day 1, Day 2, Day 3, and Day 4. (C) The cell proliferation was measured by BrdU ELISA assay after the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic or negative control on Day 4. (D) The ability to form colonies was determined by colony formation assay after the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic or negative control. (E) The cell migration was assessed by wound healing assay after the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic or negative control at 0 h and 24 h. (F) The cell adhesion ability was assessed by cell adhesion assay after the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic or negative control at 30 min and 60 min. CON, the cells in the control group were cultured without any treatment. NC, the cells in the NC group were transfected with negative control. mimic, the cells in the mimic group were transfected with miR-1298-5p mimic. *P<0.05, **P<0.001 versus CON.
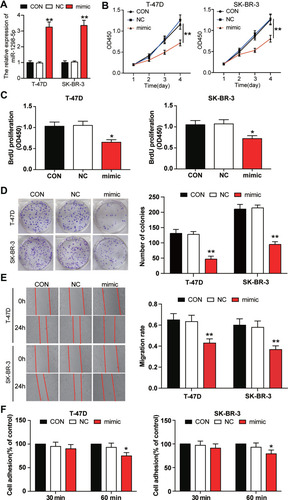
Figure 4 miR-1298-5p directly targeted CXCL11. (A) The binding site between miR-1298-5p and CXCL11 predicted by starBase was illustrated. (B) The binding site was validated using the dual luciferase reporter gene assay after the T-47D and and SK-BR-3 cells were co-transfected with miR-1298-5p mimic/NC and pGL3-CXCL11-3′UTR-WT/pGL3-CXCL11-3′UTR-Mut vectors. WT (wild-type), pGL3-CXCL11-3′UTR-WT. MUT (mutant), pGL3-CXCL11-3′UTR-Mut. **P<0.001 versus pGL3-CXCL11-3′UTR-WT+NC group. (C) The expression of CXCL11 mRNA was detected by RT-qPCR in T-47D and SK-BR-3 cells transfected with miR-1298-5p mimic or negative control. CON, the cells in the control group were cultured without any treatment. NC, the cells in the NC group were transfected with negative control. mimic, the cells in the mimic group were transfected with miR-1298-5p mimic. **P<0.001 versus CON. (D) The expression of CXCL11 protein was detected by Western blotting in T-47D and SK-BR-3 cells transfected with miR-1298-5p mimic or negative control. (E) The protein level of CXCL11 secreted into the culture medium was detected by CXCL11 ELISA assay in T-47D and SK-BR-3 cells transfected with miR-1298-5p mimic or negative control. CON, the cells in the control group were cultured without any treatment. NC, the cells in the NC group were transfected with negative control. mimic, the cells in the mimic group were transfected with miR-1298-5p mimic. *P<0.05, **P<0.001 versus CON.
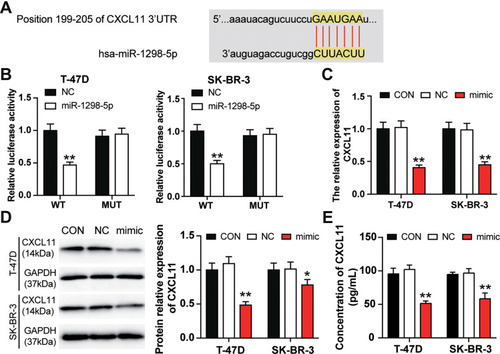
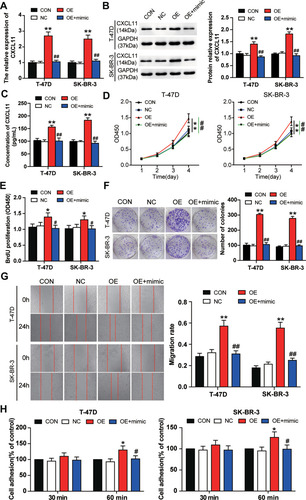
Figure 5 miR-1298-5p reversed the promotion effect of CXCL11 on BC cell phenotypes. (A) The expression of CXCL11 mRNA was measured by RT-qPCR in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (B) The protein expression of CXCL11 was measured by Western blotting in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (C) The protein level of CXCL11 secreted into the culture medium was detected by CXCL11 ELISA assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (D) The cell viability was measured by CCK-8 assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic on Day 1, Day 2, Day 3, and Day 4. (E) The cell proliferation was measured by BrdU ELISA assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (F) The ability to form colonies was determined by colony formation assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (G) The cell migration was assessed by wound healing assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. (H) The cell adhesion ability was assessed by cell adhesion assay in T-47D and SK-BR-3 cells transfected with negative control, CXCL11 overexpression vectors, and CXCL11 overexpression vectors+miR-1298-5p mimic. CON, the cells in the control group were cultured without any treatment. NC, the cells in the NC group were transfected with negative control. OE, the cells in the OE group were transfected with CXCL11 overexpression vectors. OE+mimic, the cells in the OE+mimic group were co-transfected with CXCL11 overexpression vectors and miR-1298-5p mimic. *P<0.05, **P<0.001 versus CON group. #P<0.05, ##P<0.001 versus OE group.