Figures & data
Figure 1 Poor prognosis of ESCC patients was correlated with the high expression level of CircRIMS in ESCC tissues. ESCC and paired non-tumor tissue samples were subjected to RNA isolation and RT-qPCR to determine the differential expression of CircRIMS in ESCC, which is expressed as a heatmap plotted using Heml 1.0 software (A). With the median expression level of CircRIMS in ESCC tissues as a cutoff value, the 64 ESCC patients were grouped into high and low (n=32) level groups (B).
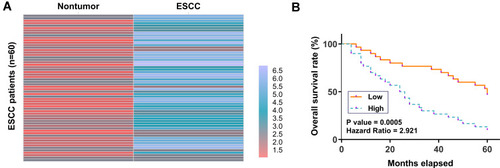
Figure 2 MiR-613 was under-expressed in ESCC and inversely correlated with CircRIMS. ESCC and paired non-tumor tissue samples were subjected to RNA isolation and RT-qPCR to determine the differential expression of miR-613 in ESCC, which is expressed as a heatmap plotted using Heml 1.0 software (A). Pearson’s correlation coefficient analysis was performed to analyze the correlations between CircRIMS and miR-613 ESCC tissue samples (B) and non-tumor tissues (C).
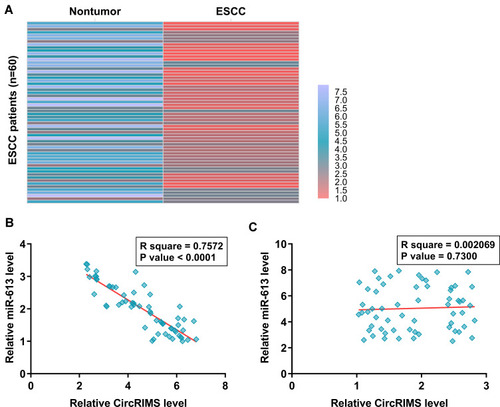
Figure 3 CircRIMS directly targets miR-613. Luciferase activities were used to investigate the targeting of circNELL2 and miR-127-5p (A). FISH experiment using the probe of miR-613 and CircRIMS were carried out to evaluate the co-location of them (B). *p<0.05.
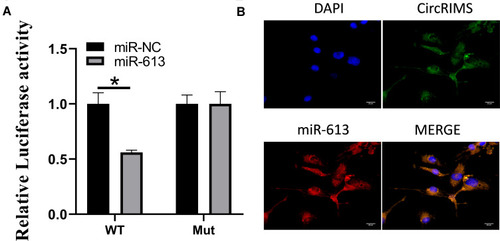
Figure 4 CircRIMS overexpression decreased the expression of miR-613 in KYSE450 cells through methylation. To study the crosstalk between CircRIMS and miR-613, KYSE450 cells were transfected with either CircRIMS expression vector or miR-613 mimic, followed by the confirmation of the overexpression of CircRIMS and miR-613 by RT-qPCR every 24h until 96h (A). The effects of CircRIMS overexpression on the expression of miR-613 (B), and the effects of miR-613 overexpression on the expression of CircRIMS (C) were analyze by RT-qPCR. MSP was performed to analyze the role of CircRIMS in regulating the methylation of miR-613 gene (D). The expression of DNMT1 (E) and Ki67 (F) was detected by Western blot assay. M, methylated PCR products; U, un-methylated PCR products; *p<0.05.
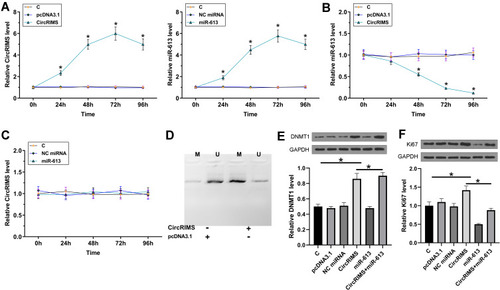
Figure 5 CircRIMS overexpression reduced the inhibitory effects of miR-613 overexpression on cell proliferation. The role of CircRIMS and miR-613 in regulating the proliferation of KYSE450 cells was analyzed by BrdU assay. Experiments were repeated 3 times and mean±values of three biological replicates were present an compared. *p<0.05.
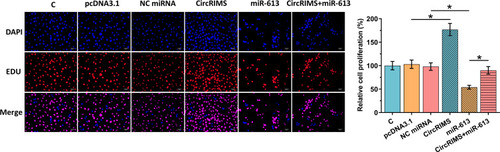
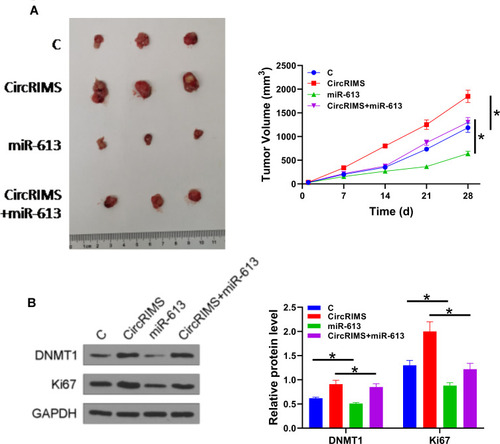
Figure 6 CircLPAR3 increased the tumor growth of ESCC by downregulating miR-613 through methylation. A xenograft tumour model was established by subcutaneously injecting transfected KYSE450 cells. Tumour image was shown in each group (A). The expression of DNMT1 and Ki67 was detected by Western blot assay (B). Experiments were repeated 3 times and mean±values of three biological replicates were present an compared. *P < 0.05.