Figures & data
Figure 1 Expression of TINCR and miR-761 in serum of early TNBC patients and early BC patients. (A) The results of RT-PCR showed that the expression of TINCR in the serum of early TNBC patients was significantly higher than that of early BC patients. (B) The results of RT-PCR showed that the expression of miR-761 in the serum of early TNBC patients was significantly higher than that of early BC patients. (C) Pearson correlation coefficient analysis showed that there was a significant positive correlation between TINCR and miR-761 in serum of early TNBC patients (r=0.646, P<0.001). (D) ROC curve analyzed the value of serum TINCR and miR-761 in differentiating early TNBC patients from early BC patients.

Figure 2 Regulatory effects of TINCR on the metastatic potential of TNBC cells. (A) The results of RT-PCR showed that TINCR was generally up-regulated in TNBC cells, and its expression level was higher in BT-549 and SUM159PT cells. (B) The results of RT-PCR showed that transfection of TINCR and si-TINCR could up-regulate and down-regulate TINCR, respectively. (C and D) Transwell showed that TINCR could regulate the migration level of BT-549 and SUM159PT cells. (E and F) Transwell showed that TINCR could regulate the invasion level of BT-549 and SUM159PT cells. (G and H) Western blot analysis showed that up-regulation of TINCR significantly promoted EMT, while down-regulation of TINCR significantly inhibited EMT, as well as its protein profiling.
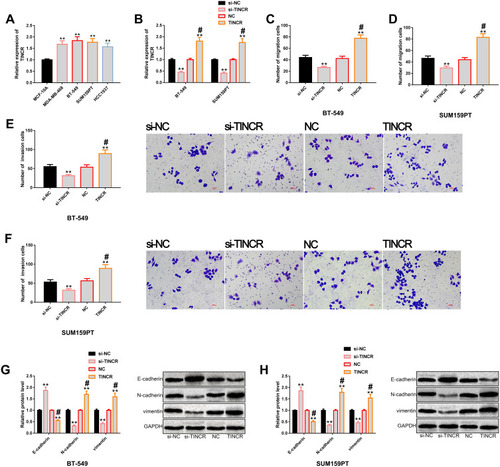
Figure 3 miR-761 regulates the metastatic potential of TNBC cells. (A) The results of RT-PCR showed that miR-761 was generally up-regulated in TNBC cells, and its expression level was higher in BT-549 and SUM159PT cells. (B) The results of RT-PCR showed that transfection of miR-761 and anti-miR-761 realized up-regulation and down-regulation of miR-761, respectively. (C and D) Transwell showed that miR-761 could regulate the migration level of BT-549 and SUM159PT cells. (E and F) Transwell showed that miR-761 could regulate the invasion level of BT-549 and SUM159PT cells. (G and H) Western blot analysis showed that up-regulation of miR-761 significantly promoted EMT, while down-regulation of miR-761 significantly inhibited EMT, as well as its protein profiling.
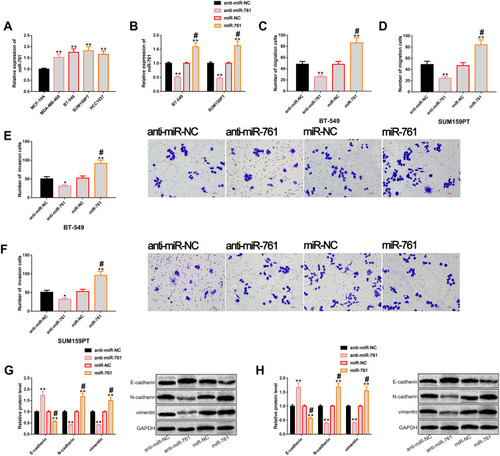
Figure 4 Positive regulation of TINCR on miR-761. (A) Binding sites of TINCR and miR-761 were found in miRDB. (B and C) The results of dual luciferase gene report showed that miR-761 in BT-549 and SUM159PT cells only affected TINCR-Wt, but not TINCR-Mut. (D) RNA pull-down indicated that Bio-TINCR could significantly recruit miR-761. (E and F) The results of RT-PCR showed that TINCR in BT-549 and SUM159PT cells could positively regulate the expression level of miR-761.
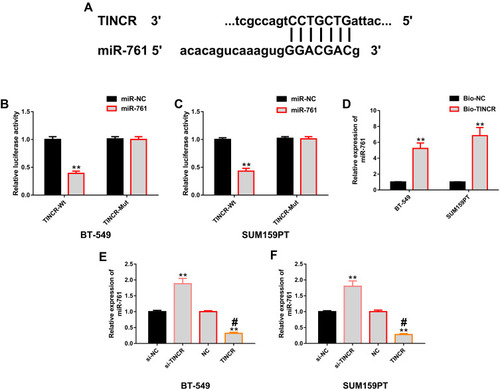
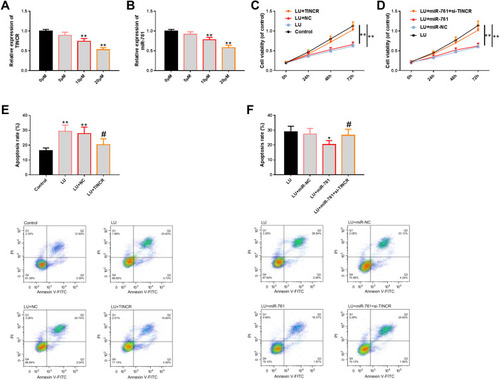
Figure 5 Effect of TINCR-miR-761 molecular module on the anti-TNBC activity of LU. (A and B) The results of RT-PCR showed that LU could affect the expression of TINCR and miR-761 in a dose-dependent manner. (C and D) The results of CCK-8 showed that the molecular module of TINCR-miR-761 could at least partially affect the anti-proliferation level of LU. (E and F) FCM showed that the molecular module of TINCR-miR-761 could at least partially affect the apoptosis-inducing level of LU, as well as its diagram.