Figures & data
Figure 1 Microarray assay result of GSE126094.
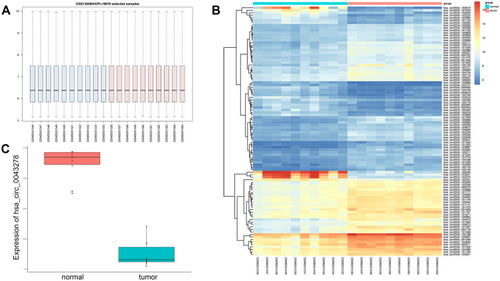
Figure 2 Sanger sequencing result of hsa_circ_0043278.
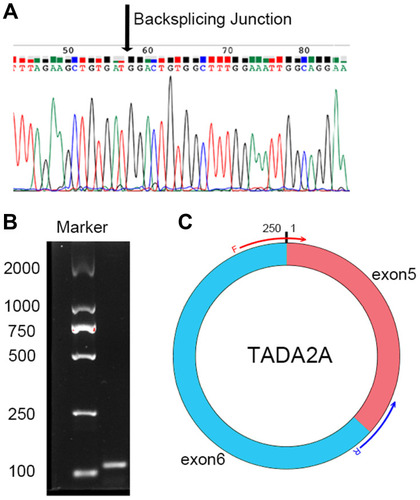
Figure 3 The expression level of hsa_circ_0043278 in colorectal cancer tissues and cell lines.
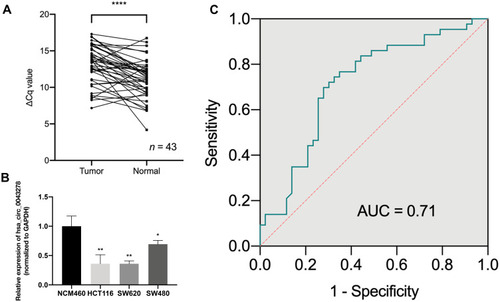
Table 1 The Association Between Hsa_circ_0043278 and Clinicopathological Factors in CRC Patients
Figure 4 Hsa_circ_0043278 inhibits colorectal cancer cell proliferation.
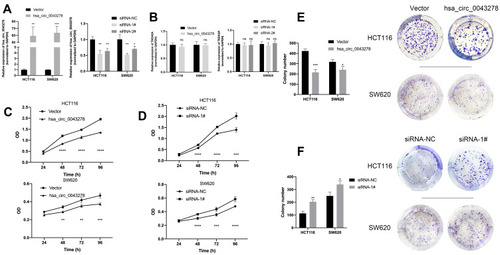
Figure 5 Hsa_circ_0043278 promotes colorectal cancer cell apoptosis.
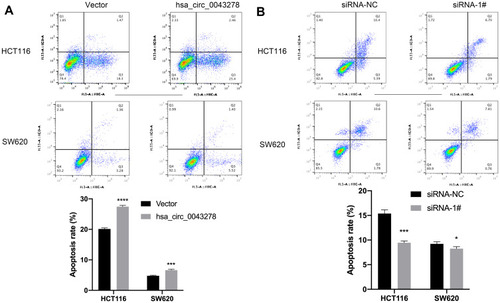
Figure 6 Hsa_circ_0043278 affects colorectal cancer cell cycle.
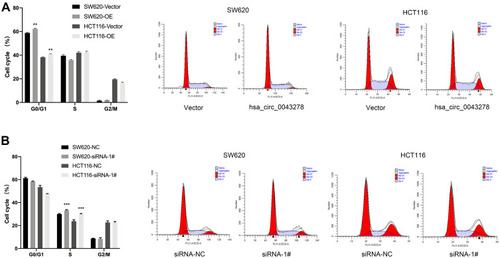
Figure 7 Hsa_circ_0043278 inhibits colorectal cancer cell migration.
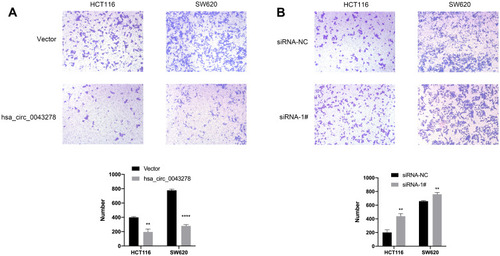
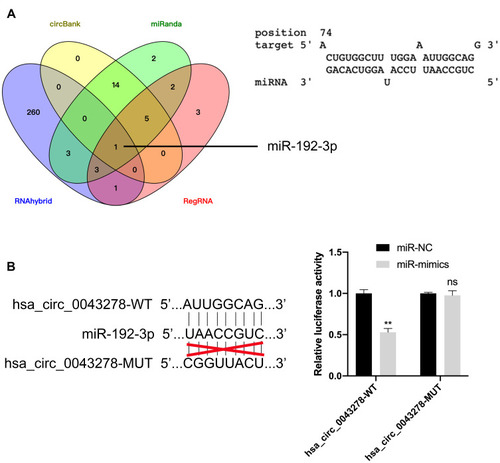
Figure 8 Hsa_circ_0043278 sponges with miR-192-3p.