Figures & data
Figure 1 HCG18 is overexpressed in ccRCC and knockdown of HCG18 represses the tumorigenesis of ccRCC. (A and B) RT-qPCR assay was applied to detect the expression of HCG18 in ccRCC tissues (n=32) and cell lines. (C) The ROC curve of serum HCG18 for diagnosing ccRCC. (D) Kaplan–Meier method was used to assess the association between HCG18 expression and overall survival rate in ccRCC patients. (E) RT-qPCR assay was applied to detect the transfection efficiency of shNC and shHCG18 in Caki-1 and 786-O cells. (F) CCK-8 assay was used to measure cell proliferation in Caki-1 and 786-O cells after knocking down HCG18. (G and H) Transwell assay was performed to assess cell migration and invasion in Caki-1 and 786-O cells transfected with shHCG18. (I and J) In vivo experiments the tumor growth rate in mice of ccRCC cells transfected with shHCG18. *p < 0.05.
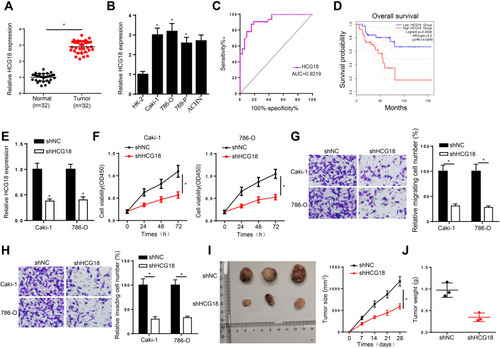
Figure 2 HCG18 is a molecular sponge for miR-152-3p in ccRCC. (A) Binding sequences between HCG18 and miR-152-3p were predicted by starBase website. (B) Luciferase reporter assay was adopted to verify the binding ability between HCG18 and miR-152-3p in 293T cells. (C and D) RIP assay was used to analyze enrichment of HCG18 and miR-152-3p in Caki-1 and 786-O cells of anti-Ago2 group compared with anti-IgG group. (E) RT-qPCR assay was performed to measure the expression of miR-152-3p in ccRCC tissues. (F) Pearson’s correlation analysis showed the correlation between HCG18 and miR-152-3p in ccRCC tissues. (G) RT-qPCR analysis was applied to detect miR-152-3p expression in Caki-1 and 786-O cells transfected with shHCG18. *p < 0.05.
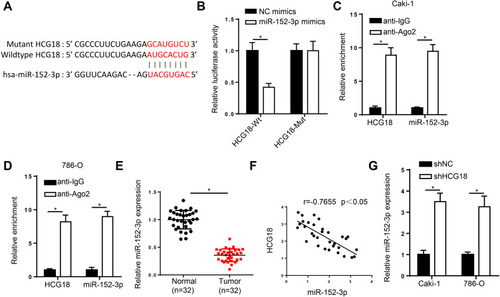
Figure 3 HCG18 sponges miR-152-3p and positively modulates RAB14 expression in ccRCC. (A) Binding sequences between miR-152-3p and RAB14 were predicted by starBase website. (B) RT-qPCR assay showed the expression of RAB14 in ccRCC tissues. (C) RT-qPCR assay was applied to assess the expression of RAB14 in Caki-1 and 786-O cells transfected with shHCG18. (D and E) Pearson’s correlation analysis showed the correlation between RAB14 and miR-152-3p or HCG18 in ccRCC tissues. (F) RT-qPCR assay was applied to assess the expression of miR-152-3p in Caki-1 and 786-O cells transfected with miR-152-3p inhibitor. (G) RT-qPCR assay showed RAB14 expression in Caki-1 and 786-O cells transfected with shNC, shHCG18, and shHCG18+miR-152-3p inhibitor. *p < 0.05.
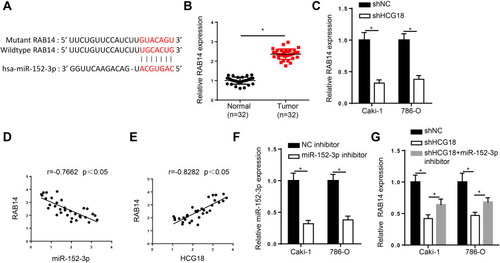
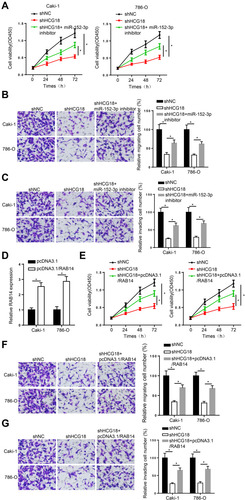
Figure 4 HCG18 plays an oncogenic role in ccRCC by regulating the miR-152-3p/RAB14 axis. (A–C) CCK-8 and transwell assays were used to test the proliferation, migration and invasion abilities in Caki-1 and 786-O cells transfected with shNC, shHCG18, and shHCG18+miR-152-3p inhibitor. (D) RT-qPCR assay showed the expression of RAB14 in Caki-1 and 786-O cells transfected with pcDNA3.1/RAB14. (E–G) CCK-8 and transwell assays were used to measure the proliferation, migration and invasion abilities in Caki-1 and 786-O cells transfected with shNC, shHCG18, and shHCG18+pcDNA3.1/RAB14. *p < 0.05.