Figures & data
Table 1 Patient Characteristics of Stool Samples Used in This Study
Table 2 Other Clinicopathological Characteristics of Ade
Table 3 Primers and Probes Used in This Study
Figure 1 SDC2 promoter methylation level in subgroups of patients with CRC, stratified based on gender, age and other criteria (UALCAN). (A) Boxplot showing relative promoter methylation level of SDC2 in normal and CRC samples. (B) Boxplot showing relative promoter methylation level of SDC2 in normal individuals of either gender or male or female CRC patients. (C) Boxplot showing relative promoter methylation level of SDC2 in normal individuals of any age or in CRC patients aged 21–40, 41–60, 61–80, or 81–100 yr. (D) Boxplot showing relative promoter methylation level of SDC2 in normal individuals of either race or in CRC patients of Caucasian, African-American or Asian. (E) Boxplot showing relative promoter methylation level of SDC2 in normal individuals of either weight or in CRC patients with normal weight, extreme weight, obese or overweight. (F) Boxplot showing relative promoter methylation level of SDC2 in normal individuals or in CRC patients in stages 1, 2, 3 or 4 tumors. (G) Boxplot showing relative promoter level of SDC2 in normal individuals of either histology or adenocarcinoma or mucinous adenocarcinoma CRC patients. (H) Boxplot showing relative promoter methylation level of SDC2 in normal individuals of any nodal metastasis status or in CRC patients in N0, N1, N2. ***p< 0.001.
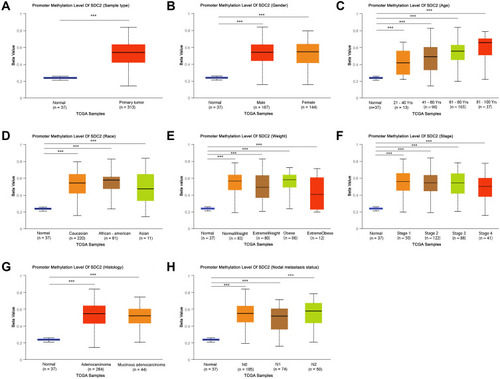
Figure 2 TFPI2 promoter methylation level in subgroups of patients with CRC, stratified based on gender, age and other criteria (UALCAN). (A) Boxplot showing relative promoter methylation level of TFPI2 in normal and CRC samples. (B) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals of either gender or male or female CRC patients. (C) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals of any age or in CRC patients aged 21–40, 41–60, 61–80, or 81–100 yr. (D) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals of either race or in CRC patients of Caucasian, African-American or Asian. (E) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals of either weight or in CRC patients with normal weight, extreme weight, obese or overweight. (F) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals or in CRC patients in stages 1, 2, 3 or 4 tumors. (G) Boxplot showing relative promoter level of TFPI2 in normal individuals of either histology or adenocarcinoma or mucinous adenocarcinoma CRC patients. (H) Boxplot showing relative promoter methylation level of TFPI2 in normal individuals of any nodal metastasis status or in CRC patients in N0, N1, N2. ***p < 0.001.
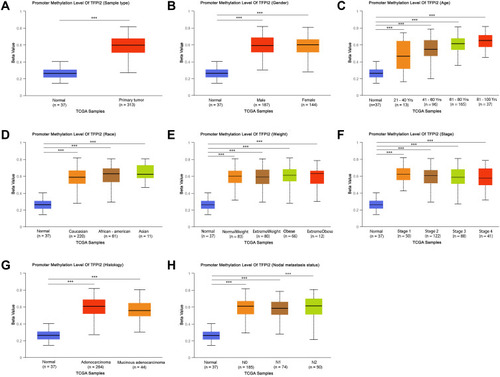
Figure 3 Heatmap for ACTB, SDC2 and TFPI2 in CRC. (A) The heatmap for ACTB in TCGA-CRC using MethSurv. (B) The heatmap for SDC2 in TCGA-CRC using MethSurv. (C) The heatmap for TFPI2 in TCGA-CRC using MethSurv.

Figure 4 Methylation levels of CpG citesof ACTB, SDC2 and TFPI2 in CRC and Normal. (A) Methylation levels throughout the region around ACTB gene between normal tissues and CRC tumors are compared. (B) Methylation levels throughout the region around SDC2 gene between normal tissues and CRC tumors are compared. (C) Methylation levels throughout the region around TFPI2 gene between normal tissues and CRC tumors are compared. Green probes indicate CpG islands.
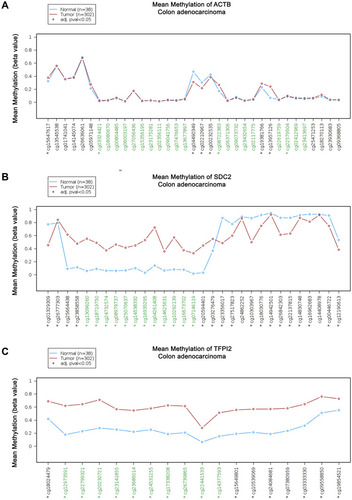
Figure 5 Kaplan-Meier survival curves comparing the high and low methylation levels of SDC2 and TFPI2 in CRC patients (UCSC-XENA). (A) The overall survival curve for CRC patients with high or low methylation level of SDC2. (B) The disease-specific survival curve for CRC patients with high or low methylation level of SDC2. (C) The overall survival curve for CRC patients with high or low methylation level of TFPI2. (D) The disease survival curve for CRC patients with high or low methylation level of TFPI2.
Figure 6 Genes differentially expressed in correlation with SDC2 methylation in CRC (LinkedOmics). (A) A Pearson test was used to analyze correlations between SDC2 methylation and genes differentially expressed in CRC. (B and C) Heat maps showing genes positively and negatively correlated with SDC2 methylation in CRC (TOP 50). Red indicates positively correlated genes and green indicates negatively correlated genes.

Figure 7 Genes differentially expressed in correlation with TFPI2 methylation in CRC (LinkedOmics). (A) A Pearson test was used to analyze correlations between TFPI2 methylation and genes differentially expressed in CRC. (B and C) Heat maps showing genes positively and negatively correlated with TFPI2 methylation in CRC (TOP 50). Red indicates positively correlated genes and green indicates negatively correlated genes.

Figure 8 Significantly enriched GO annotations and KEGG pathways about SDC2 methylation in CRC. (A) Cellular components. (B) Biological processes. (C) Molecular functions. (D) KEGG pathway analysis.
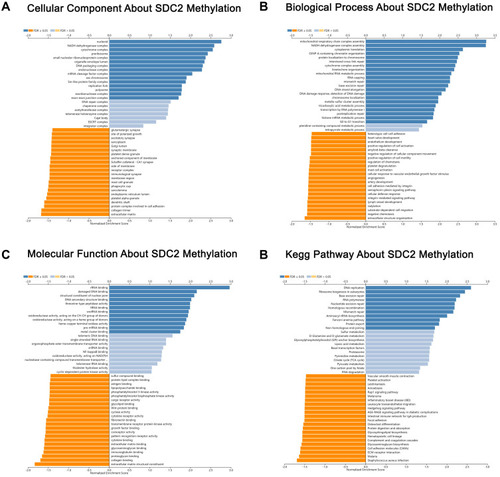
Figure 9 Significantly enriched GO annotations and KEGG pathways about TFPI2 methylation in CRC. (A) Cellular components. (B) Biological processes. (C) Molecular functions. (D) KEGG pathway analysis.
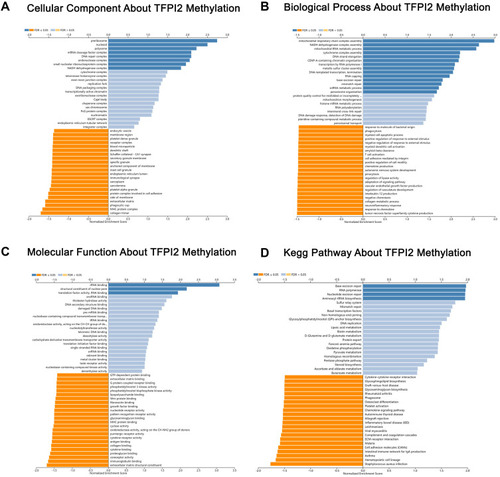
Table 4 Positive Detection Rates and Youden Indexes for Integration of Methylated SDC2 and TFPI2 in Detecting Normal, Ade and CRC in Stool Samples. N/A, Not Applicable
Table 5 Correlation Between Methylation Detection Rate and the Clinicopathologic Features in CRC Stool Samples
Table 6 Correlation Between Methylation Detection Rate and the Clinicopathologic Features in Ade Stool Samples
Table 7 Comparison of Methylation Detection of CRC, Gastric Cancer and Other Gastrointestinal Tumors
Table 8 Comparison of Integrated Detection of Methylated SDC2 and TFPI2 to Other Screening Methods in CRC
