Figures & data
Figure 1 AVL9 expression in CRC and normal tissues detected using bioinformatics methods.
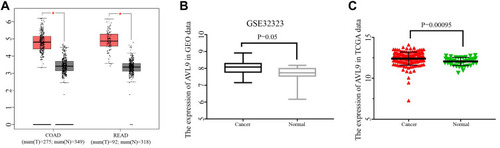
Figure 2 Analysis of the relationship between AVL9 expression and prognosis using bioinformatics methods.
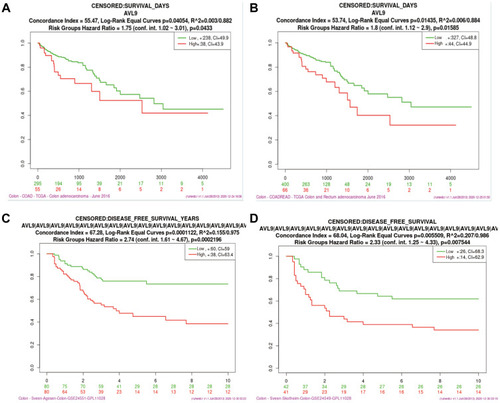
Figure 3 Bioinformatics analysis of genes related to AVL9 expression.
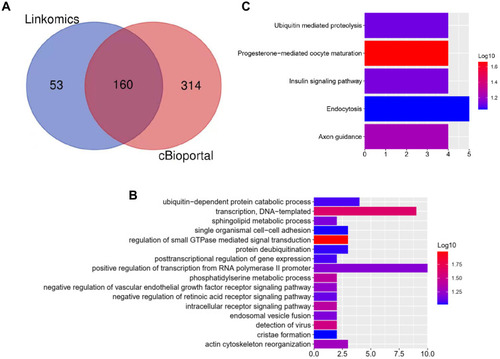
Figure 4 PPI network analysis of AVL9 expression-related genes and key gene predictions.
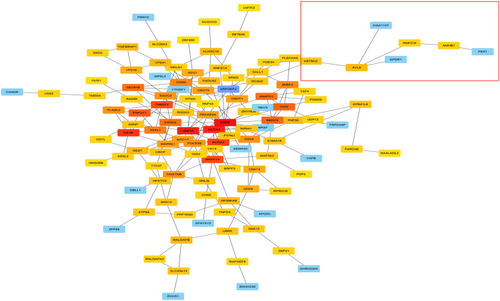
Table 1 AVL9 Expression and Clinicopathological Features in Patients with Colorectal Cancer
Table 2 Univariate and Multivariate Analyses of Clinical Pathological Factors of OS in 50 Patients with CRC
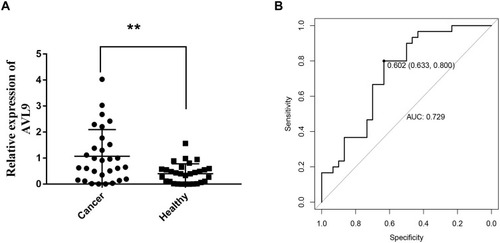
Figure 5 Expression level of AVL9 in CRC tissues.
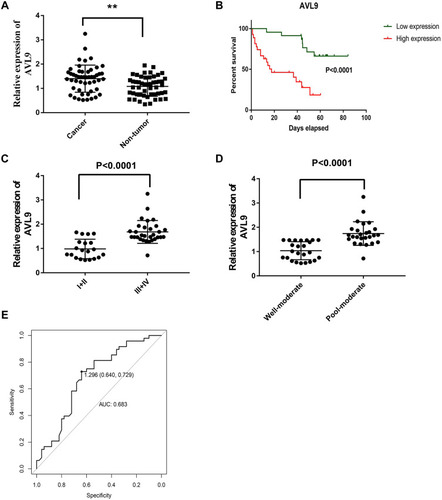
Figure 6 Diagnostic utility of circulating AVL9 was tested in 30 CRC patients and 30 healthy subjects.