Figures & data
Table 1 Correlation Between LncRNA HOXA-AS2 Expression and the Clinical Characteristics of OSCC
Figure 1 The expression of lncRNA HOXA-AS2 and miR-567 in OSCC Clinical Tissue Samples and Cell Lines. (A) The expression levels of lncRNA HOXA-AS2 in OSCC clinical tissue samples and adjacent normal tissue samples were detected by qRT-PCR. (B) The expression of miR-567 in OSCC clinical tissue samples and adjacent normal tissue samples was detected by qRT-PCR. (C) The relationship between lncRNA HOXA-AS2 and miR-567 expression in OSCC samples was analyzed using the GraphPad Prism software. (D) The expression levels of lncRNA HOXA-AS2 in four OSCC cell lines and the NHOK normal oral cell line were detected by qRT-PCR. (E) The expression of miR-567 in three OSCC cell lines and the NHOK normal oral cell line was detected by qRT-PCR. *Means P <0.05, and **Means P<0.01.
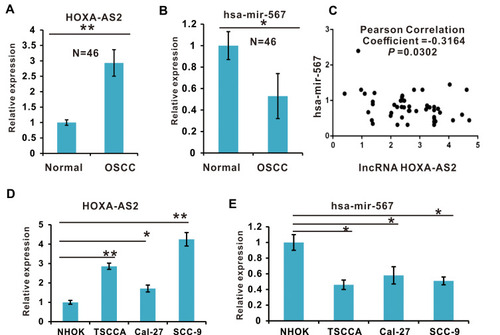
Figure 2 LncRNA HOXA-AS2 negatively regulated miR-567 expression by direct binding. (A) The putative binding sites between lncRNA HOXA-AS2 and miR-567. (B) The construction of a luciferase reporter expressing lncRNA HOXA-AS2. (C) The luciferase activities of lncRNA HOXA-AS2-WT, lncRNA HOXA-AS2-Mut1 or lncRNA HOXA-AS2-Mut2 in TSCCA OSCC cells with or without miR-567 co-transfection were evaluated by a luciferase reporter assay. (D) The relative expression of miR-567 in TSCCA OSCC cells transfected with lncRNA HOXA-AS2-WT, lncRNA HOXA-AS2-Mut1, lncRNA HOXA-AS2-Mut2 or their corresponding vector control. **Means P<0.01.
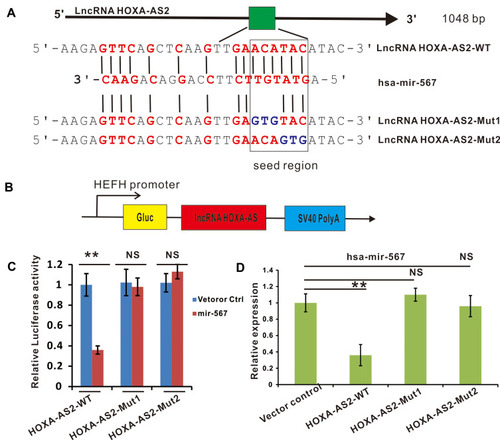
Figure 3 Cell proliferation was promoted in lncRNA HOXA-AS2 overexpressing OSCC cell lines. (A) The relative expression of lncRNA HOXA-AS2 in TSCCA and SCC-9 cells transfected with lncRNA HOXA-AS2 was examined by qRT-PCR. (B) The relative expression of miR-567 in TSCCA and SCC-9 cells with lncRNA HOXA-AS2 overexpression. (C and D) The proliferation of TSCCA and SCC-9 cells with lncRNA HOXA-AS2 overexpression was evaluated by the WST-1 assay and crystal violet staining.(E) Cell cycle were determined in TSCCA and SCC-9 cells treated with or without lncRNA HOXA-AS2 overexpression. *Means P <0.05, **Means P<0.01, ***Means P<0.001.
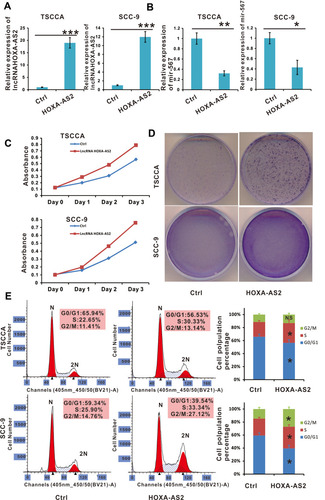
Figure 4 Cell proliferation was promoted in OSCC cell lines with lncRNA HOXA-AS2 silenced by siRNA. (A) The relative expression of lncRNA HOXA-AS2 in TSCCA and SCC-9 cells with lncRNA HOXA-AS2 silenced was examined by qRT-PCR. (B) The relative expression of miR-567 in TSCCA and SCC-9 cells with LncRNA HOXA-AS2 silenced. (C and D) The proliferation of TSCCA and SCC-9 cells with lncRNA HOXA-AS2 silenced was evaluated by the WST-1 assay and crystal violet staining. (E) Cell cycle analysis was determined in TSCCA and SCC-9 cells treated with or without silencing lncRNA HOXA-AS2. *Means P <0.05, **Means P<0.01.
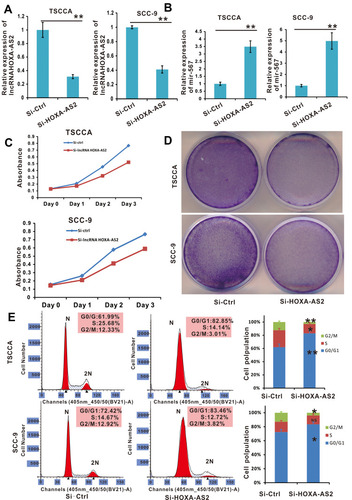
Figure 5 CDK8 is a downstream target of miR-567, and is involved in the regulation of OSCC cell proliferation. (A) A bioinformatic analysis indicated the presence of a putative binding site and a corresponding mutant region for CDK8 within miR-567. (B) The construction of a luciferase reporter that expresses CDK8. (C) The luciferase activities in CDK8-WT or CDK8-Mut and miR-567 or miR-Ctrl co-transfected OSCC cells were evaluated by a luciferase reporter assay. (D) The relative expression of CDK8 in miR-567-mimic, Mimic-Ctrl and vehicle-treated OSCC cells. (E) The protein level of CDK8 in miR-567-mimic, Mimic-Ctrl and vehicle-treated OSCC cells (GAPDH was used as a loading control) (F and G) The proliferation of miR-567-mimic, miR-567-inhibitor and their corresponding control-treated OSCC cells was evaluated by the WST-1 assay and crystal violet staining. *Means P <0.05.
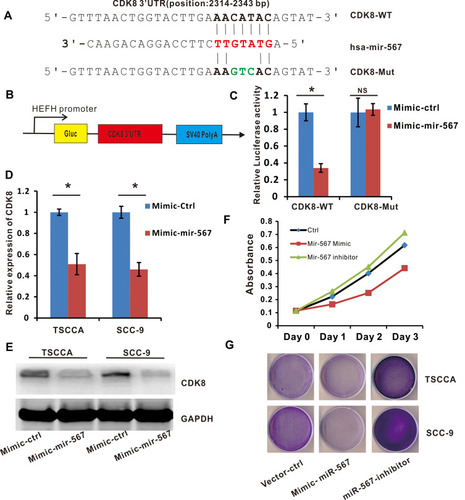
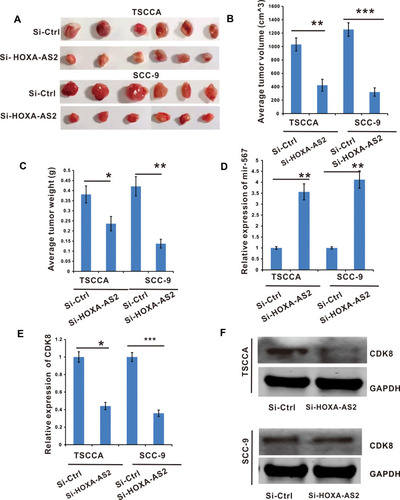
Figure 6 Silencing of lncRNA HOXA-AS2 inhibited OSCC tumor growth by releasing miR-567 to inhibit the expression of CDK8. TSCCA and SCC-9 cells with si-LncRNA HOXA-AS2 or its negative control were injected into nude mice to establish xenograft OSCC tumor models. (A) Images of tumor nodules excised at the end of the study. (B and C) The tumor volume and weight were analyzed. (D) The miR-567 expression in tumor tissues. (E) The CDK8 mRNA expression in the node mice tumor tissues were determined by RT-qPCR. (F) The CDK8 protein level in the node mice tumor tissues were determined by Western Blot. *Means P <0.05, **Means P<0.01, and ***Means P<0.001.