Figures & data
Figure 1 The expression profile comparison of lncRNAs and mRNAs between peritoneal metastasis lesion and primary foci. (A) The lncRNAs expression variation between peritoneal metastasis lesion and primary foci. (B) The mRNAs expression variation between peritoneal metastasis lesion and primary foci. The red and green dots in (A) and (B) indicated the expression levels of lncRNAs and mRNAs with fold change >2. (C) The heat map of lncRNAs. (D) The heat map of mRNAs. (E) The heat map of lncRNAs with fold change >2. The lncRNAs were selected according to whether their biological function was known.
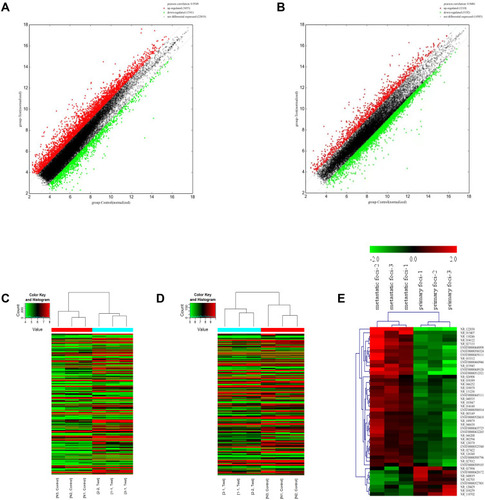
Figure 2 The coding-non-coding gene co-expression network between lncRNAs and mRNAs. (A–D) The up-regulated lncRNAs and their related mRNAs. (E) The heat map of up-regulated lncRNA-related mRNAs. (F) The down-regulated lncRNAs and their related mRNAs. (G) The heat map of down-regulated lncRNA-related mRNAs. The circle represented mRNA and the triangle represented lncRNA. Red meant up-regulated gene and green mean down-regulated gene.
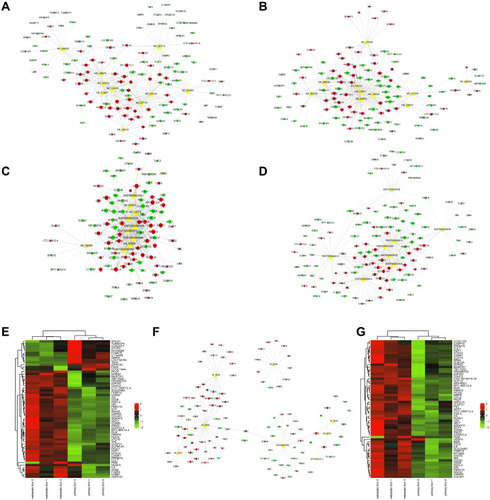
Figure 3 Gene ontology (GO) enrichment analysis. (A–C) The biological process, cellular component and molecular function of up-regulated lncRNA-related mRNA. (D–F) The biological process, cellular component and molecular function of down-regulated lncRNA-related mRNA.
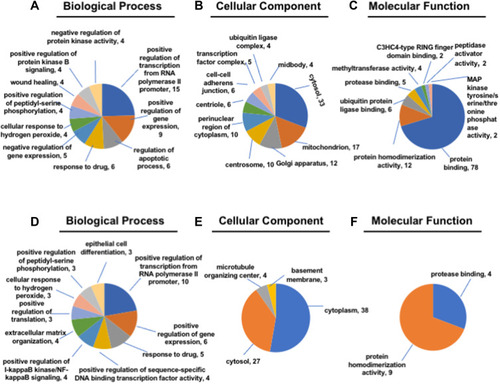
Figure 4 LncRNAs expression of gastric cancer cells after coculture with adipocytes. (A–C) The RT-qPCR was used to analyze the lncRNA expression differences of AGS with or without coculture. (D–F) The RT-qPCR was used to analyze the lncRNA expression differences of BGC823 with or without coculture.
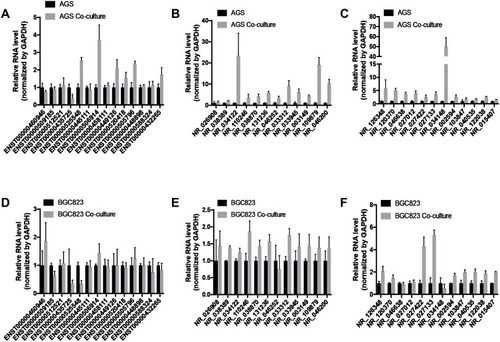
Figure 5 The function analysis of LINC00924, LINC01151 and FAM27B by cBioPortal. (A) The transcription levels of LINC00924. (B) The co-expression network of LINC00924. (C) The KEGG pathway analysis of LINC00924-related mRNAs. (D) The transcription levels of FAM27B. (E) The co-expression network of FAM27B. (F) The KEGG pathway analysis of FAM27B-related mRNAs. (G) The transcription levels of LINC01151. (H) The co-expression network of LINC01151. (I) The KEGG pathway analysis of LINC01151-related mRNAs.
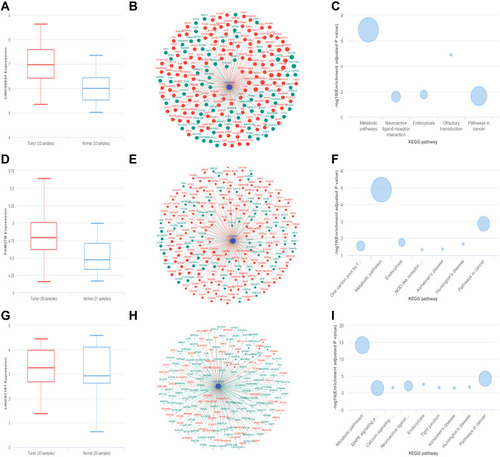
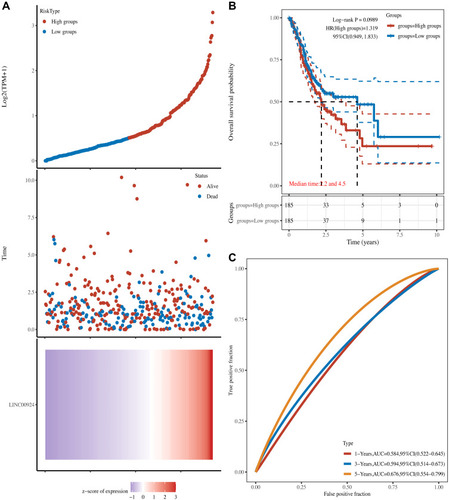
Figure 6 The univariate, multivariate analysis and survival curve of lncRNA-related mRNA derived from sequencing data of gastric cancer peritoneal metastasis in gastric cancer. (A–D) The univariate analysis. (E–H) The multivariate analysis. (I–L) Overall survival curves and different colors represented different genes.
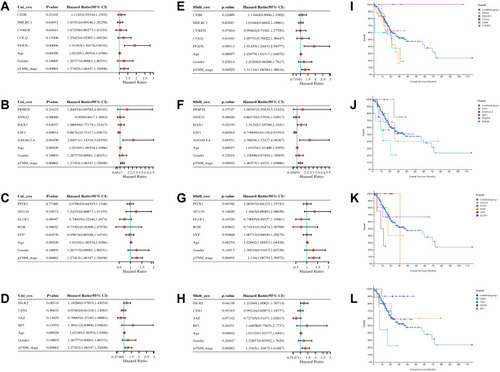
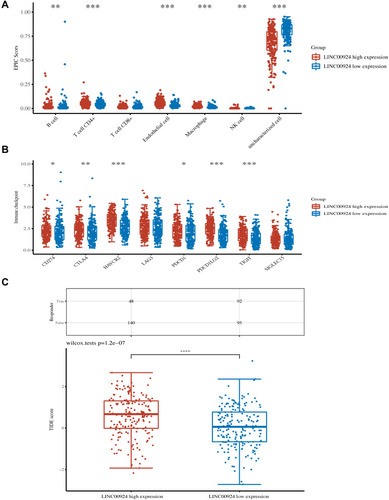
Figure 7 The relationship between LINC00924 expression and immune escape. (A) The percentage abundance of tumor-infiltrating immune cells in LINC00924 high and low expression group. (B) Immune checkpoint-related gene expression in LINC00924 high and low expression group. (C) Sample statistical table and distribution of immune response scores in LINC00924 high and low expression group in the prediction results. *P < 0.05; **P < 0.01; ***P < 0.001.