Figures & data
Figure 1 AC007207.2 is upregulated in OS tissue and cells.
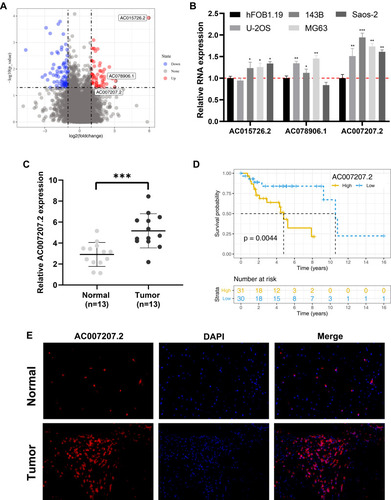
Figure 2 AC007207.2 promote cell motility ability in OS cell lines.
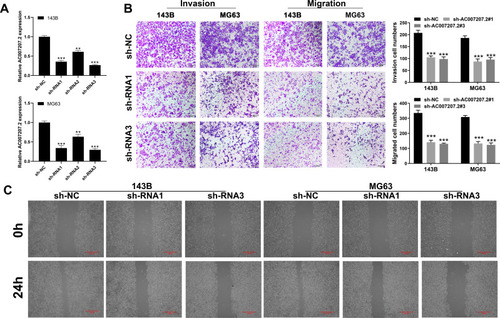
Figure 3 AC007207.2 facilitate proliferation and inhibit apoptosis in OS cell lines.
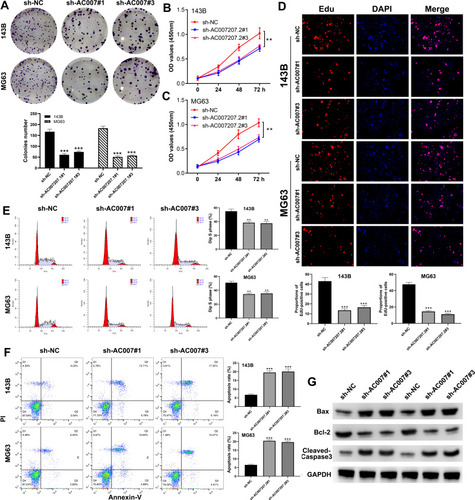
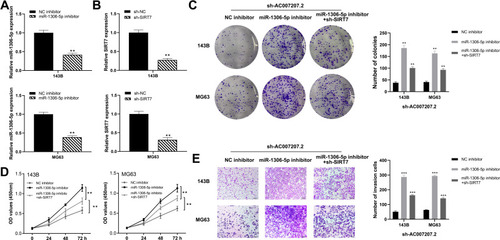
Figure 4 AC007207.2 specifically bound to miR-1306-5p.
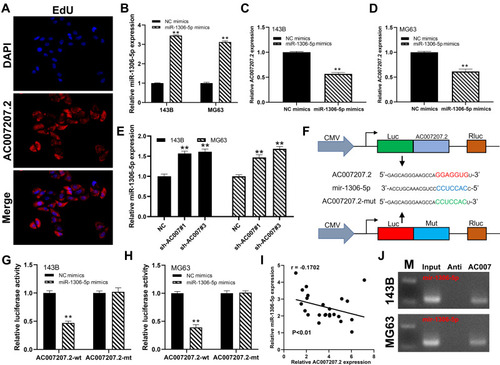
Figure 5 miR-1306-5p inhibit the proliferation and motility ability of OS cell lines.
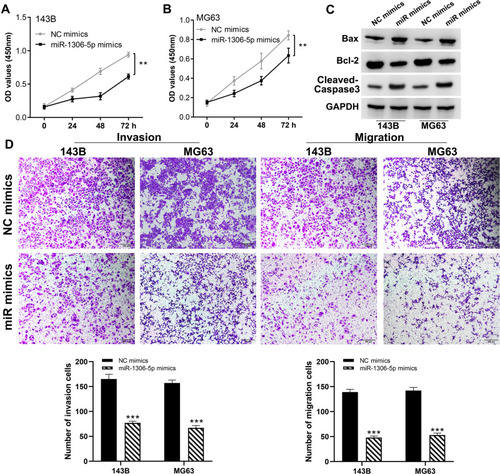
Figure 6 SIRT7 is a target gene of miR-1306-5p.
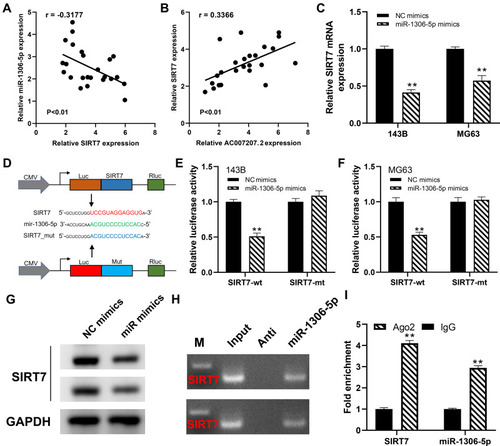
Figure 7 AC007207.2 enhances the progression of OS cells by miR-1306-5p/SIRT7 axis.