Figures & data
Figure 1 Graphical illustration showing that the regulatory mechanisms for E2Fs in CRC. For instance, E2Fs function in CRC is modulated via multiple levels including the transcriptional level (NFYB, KRT23, and ChoKa inhibitors-mediated transcription of the E2F gene), post-transcriptional regulation (E2F mRNA targeted by different miRNAs and ceRNAs), post-translational modifications (deacetylation and acetylation of E2F protein), protein–protein interaction level (phosphorylation and dephosphorylation of RB protein), and transcriptional activity level (XIAPΔRING, TRIP-Br2, cIAP1, NLK, and NPTX1 regulate the transcriptional activity of E2Fs protein). Solid arrows represent promoted effects, while dashed arrows represent inhibitory effects. Different colored lines showed different signaling pathways or targets.
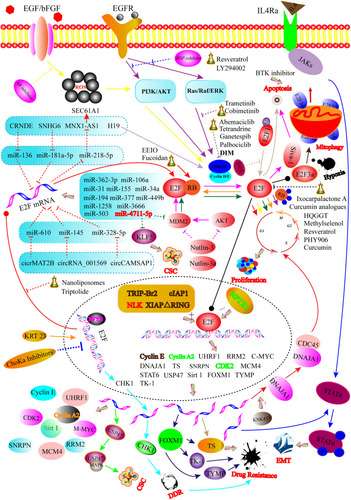
Table 1 E2Fs-Target Agents Summary in CRC
