Figures & data
Figure 1 PPI network and enrichment analysis of pyroptosis-related genes.
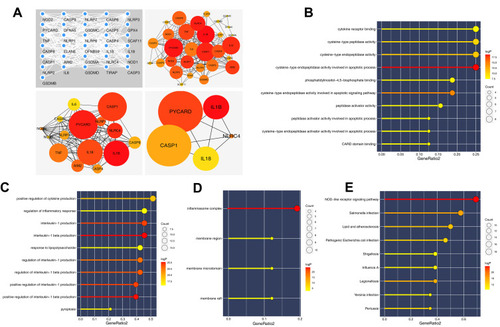
Figure 2 Construction and validation of prognosis model.
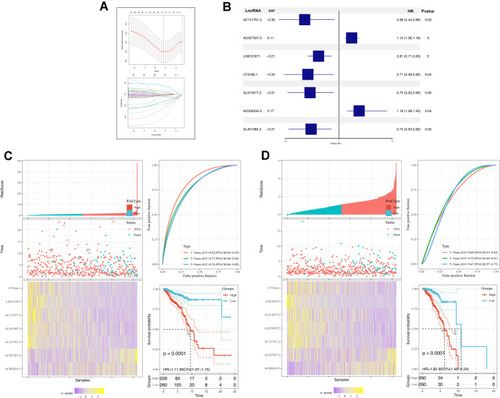
Figure 3 The independence of prognosis model and the prognosis association of model lncRNAs.
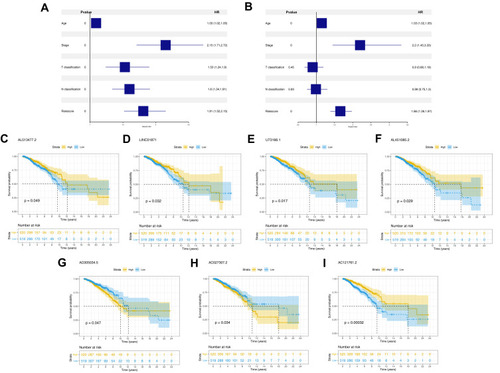
Figure 4 Clinical correlation of prognosis model and model lncRNAs.
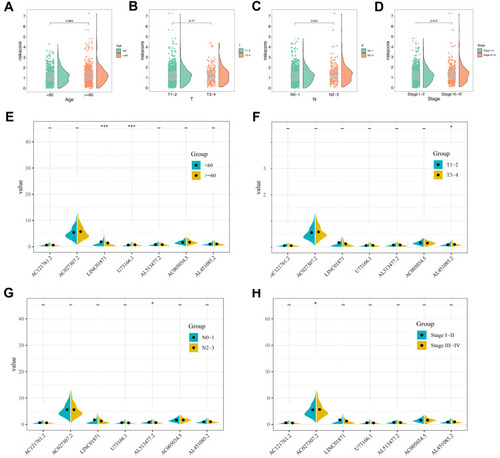
Figure 6 Immune infiltration in tumor microenvironment.
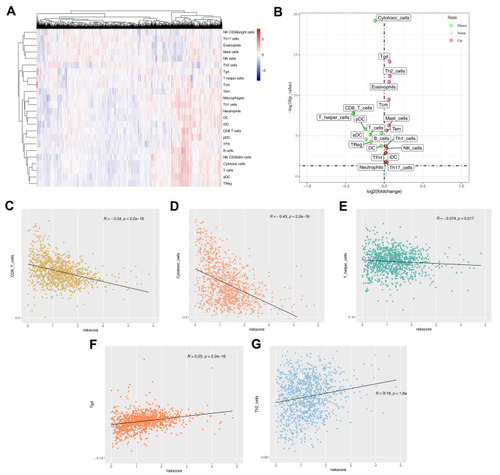
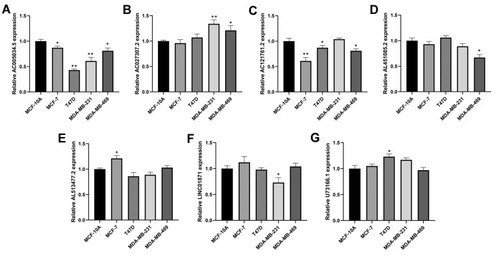
Figure 7 The qRT-PCR result of seven lncRNAs in four breast cancer and normal breast epithelial cell lines.