Figures & data
Figure 1 Large-scale identification of crotonylation sites in oral squamous cell carcinoma (OSCC) following hypoxic treatment.
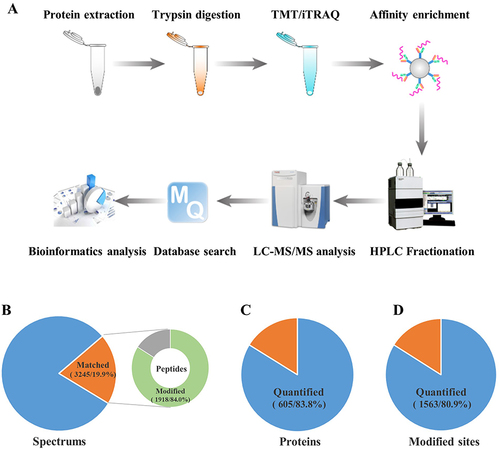
Table 1 Characteristic Sequence and Enrichment Statistical Information of Crotonylation Sites in OSCC After Hypoxic Treatment
Figure 2 Analysis of crotonylation site motifs in oral squamous cell carcinoma (OSCC).
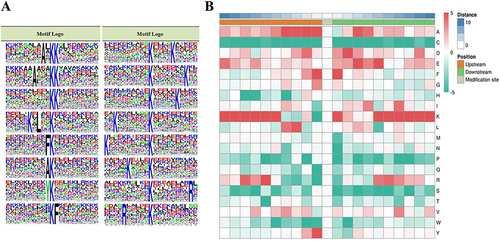
Figure 3 Analysis of differential crotonylation sites and corresponding proteins in OSCC under hypoxia conditions.
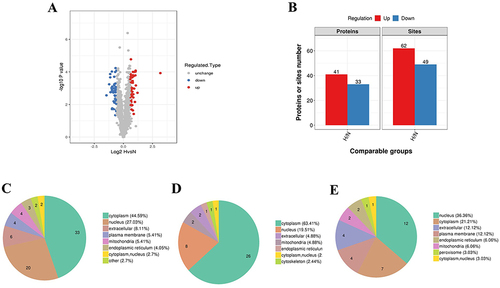
Figure 4 Functional enrichment of proteins corresponding to differential crotonylation sites in OSCC.
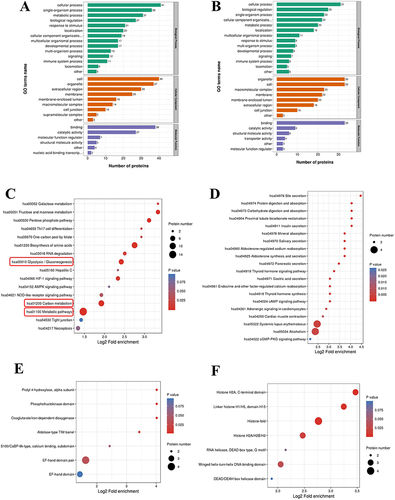
Figure 5 Functional relationships between proteins corresponding to differential crotonylation sites in OSCC.
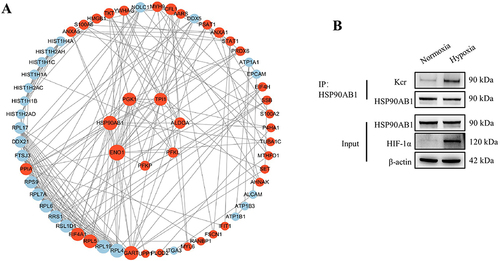
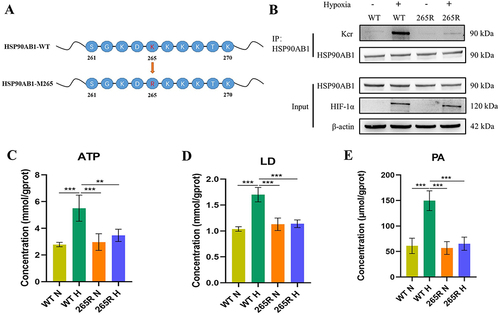
Figure 6 Crotonylation of HSP90AB1 affects its regulation of glycolysis in OSCC.