Figures & data
Table 1 Demographics Of Subjects Utilized In Histological Analysis Of MEK Pathway Activation
Figure 1 Expression of the activated MEK pathway evidenced through ERK 1/2 phosphorylation in COPD. (A–B) Increased activation of the pathway was observed in lung tissue from patients with end-stage COPD (GOLD 4) in comparison to healthy lung tissue. (C) Quantification of staining pattern in bronchial epithelia showed a significant increase (*p 0.0293 Mann–Whitney test) in pERK1/2 in severe COPD. (D) pERK1/2 staining was also observed in areas of remodeling in GOLD4 (black arrows) and (F) in alveolar macrophages in GOLD4 but not in (E) healthy lung tissue. Sections shown are representative.
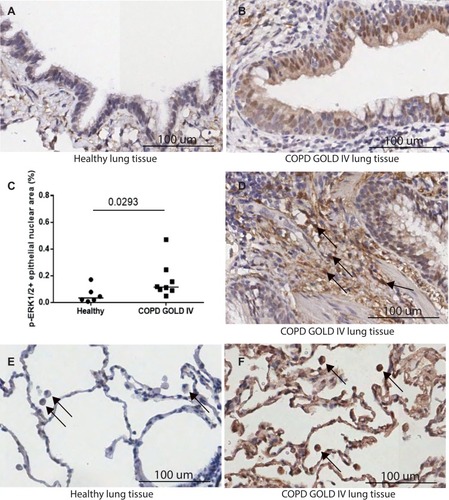
Figure 2 Transcriptomic profiling of MEK activation. (A) MEK activity in sputum from moderate and severe COPD (GSE22148) assessed as a GSVA-score using the MEK activity gene signature from Dry et al (2010).Citation17 **p<0.01 (Kruskal–Wallis test). (B) Hierarchical clustering of genes in the MEK activity gene set from Dry et al (2010)Citation17 in COPD sputum samples (GSE22148). Expression of each gene was scaled across the samples with expression going from low (blue) to high (orange). Samples are color-coded at the bottom to indicate disease severity, moderate COPD in grey and severe COPD in black. Two sub-clusters are highlighted with black boxes and denoted (i) and (ii), respectively. (C) GSVA-scores for sub-cluster (i) and (ii) gene sets from the hierarchical clustering in (B) compared across disease severity (moderate, severe) in the sputum transcriptomics dataset GSE22148. ***p<0.001 (Kruskal–Wallis test).
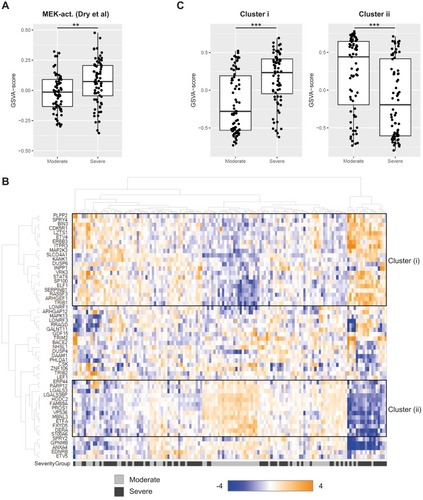
Table 2 Pathway Analysis Of MEK Activity Signature Genes. Predicted Top Upstream Regulator And Enriched Physiological Functional Terms For Genes In Cluster (i) And Cluster (ii), Respectively, Using IPA
Figure 3 Functional characterisation of MEK pathway activation in human alveolar macrophages. (A) Robust time-dependent phosphorylation of ERK1/2 was observed in human alveolar macrophages stimulated with 100ng/mL LPS (*p 0.01, ****p<0.0001 One-way Anova with Dunnett multiple comparison test). Peak phosphorylation of ERK 1/2 measured at 30min-post LPS challenge was potently inhibited by the MEK inhibitor. Data are mean ± S.E.M of 3/4 donors. (B) Potent inhibition of inflammatory cytokine release on LPS challenge in human alveolar macrophages treated with MEK or p38 kinase inhibitors. Data are mean ± S.E.M of 10/14 donors. (C) Activation of the ERK1/2 pathway through phosphorylation is observed in a concentration-dependent manner in human alveolar macrophages on treatment with p38 kinase inhibitors at 90mins post-LPS challenge (**p 0.007, ****p<0.0001 One-way Anova with Dunnett multiple comparison test). No significant activation of the p38 pathway measured through phosphorylation is observed in human alveolar macrophages on treatment with MEK kinase inhibitors at 90mins post-LPS challenge. Data are mean ± S.E.M of 4 donors.
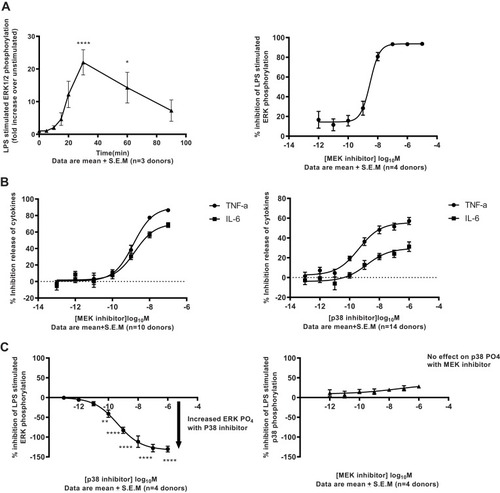
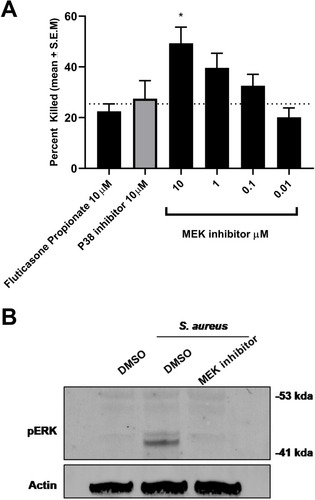
Figure 4 MEK inhibition enhances bacterial killing in RAW264.7 cells. (A) MEK inhibition results in enhanced S.aureus killing in RAW264.7 cells (*p 0.01, **p 0.002 One-way Anova with Dunnett multiple comparison test). This was not observed with p38 inhibitor or steroid Fluticasone Propionate. (B) Time-dependent activation of the MEK-pERK 1/2 pathway on S. aureus exposure in RAW264.7 cells was confirmed by Western blot analysis. Activation of the cascade was inhibited by treatment with the MEK inhibitor. Data are mean + S.E.M of 3 different experiments.
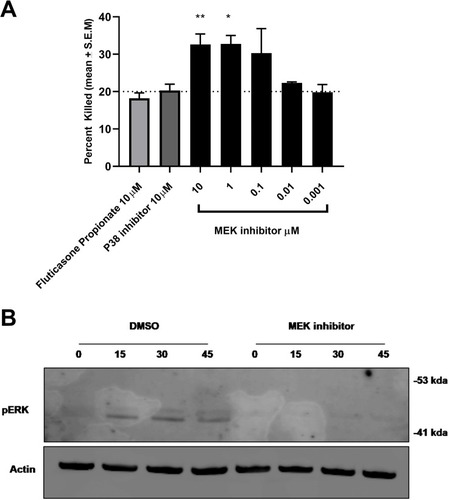
Figure 5 MEK inhibition enhances bacterial killing in human neutrophils. (A) MEK inhibition resulted in an increase in S.aureus killing in human neutrophils in a concentration-dependent manner (*p 0.01 One-way Anova with Dunnett multiple comparison test). No effect of p38 inhibitor or steroid was observed in bacterial killing in neutrophils at the concentrations tested. (B) MEK-pERK 1/2 pathway on S. aureus exposure in neutrophils was confirmed by Western blot analysis. Activation of the cascade was inhibited by treatment with the MEK inhibitor. Data are mean + S.E.M of 4 donors.