Figures & data
Figure 1 LINC00987 had low expression and let-7b-5p had high expression in LPS-induced BEAS-2B cells. (A and D) RT-qPCR was performed to detect LINC00987 and let-7b-5p expression in normal (N=33) and COPD tissues (N=29). (B and F) The expression levels of LINC00987 and let-7b-5p were determined by RT-qPCR in LPS-induced BEAS-2B cells. (C) ROC curve was used to investigate the value of LINC00987 in COPD diagnosis. (E) Spearman correlation analysis was employed to reveal the relationship between LINC00987 expression and let-7b-5p expression. **P<0.01 and ***P<0.001.
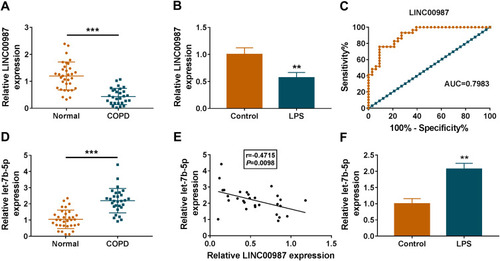
Figure 2 LINC00987 inhibited cell apoptosis, oxidative stress, inflammation and autophagy in LPS-mediated 16HBE and BEAS-2B cells. (A) The transfection efficiency of LINC00987 was detected by RT-qPCR in LPS-induced 16HBE and BEAS-2B cells. (B and C) The effects of LPS treatment and LINC00987 overexpression on LINC00987 and let-7b-5p expression were demonstrated by RT-qPCR. (D) CCK-8 assay was performed to illustrate the impacts between LPS and LINC00987 on cell viability in 16HBE and BEAS-2B cells. (E and F) Caspase3 activity and apoptosis analysis assays were conducted to present the influences between LPS and LINC00987 overexpression on caspase3 activity and cell apoptosis, respectively, in 16HBE and BEAS-2B cells. (G and H) ROS detection kit and SOD activity assays were carried out to uncover the impacts between LPS and enforced LINC00987 expression on oxidative stress in 16HBE and BEAS-2B cells. (I) Western blot assay was employed to determine the impacts of LINC00987 on the protein levels of pro-c3, cleaved-c3, SOD1, SOD2 and SOD3 in LPS-induced 16HBE and BEAS-2B cells. (J and K) ELISA kit assays were employed to exhibit the effects between LPS exposure and LINC00987 on the production of IL-6 and IL-8 in 16HBE and BEAS-2B cells. (L) The effects of LPS treatment and LINC00987 overexpression on the level of IC3-II/IC3-I and the protein expression of ATG5 and P62 were demonstrated by Western blot in 16HBE and BEAS-2B cells. *P<0.05, **P<0.01 and ***P<0.001.
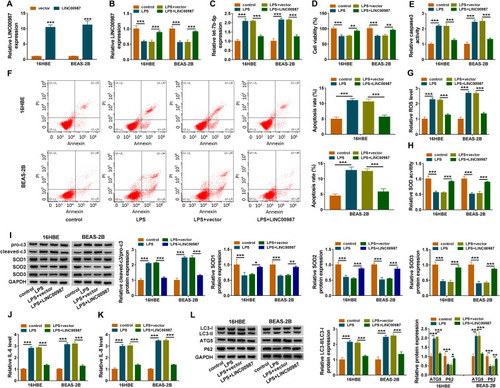
Figure 3 Let-7b-5p inhibitor attenuated LPS-induced apoptosis, oxidative stress, inflammation and autophagy in BEAS-2B cells. (A) RT-qPCR was performed to detect the transfection efficiency of anti-let-7b-5p in LPS-induced BEAS-2B cells. (B and C) The impact of anti-let-7b-5p on LINC00987 and let-7b-5p expression after LPS treatment was determined by RT-qPCR in BEAS-2B cells. (D) CCK-8 assay was carried out to illustrate the impacts between LPS and let-7b-5p absence on cell viability in BEAS-2B cells. (E and F) Caspase3 activity and apoptosis analysis assays were employed to reveal the influences between LPS exposure and let-7b-5p deletion on caspase3 activity and cell apoptosis, respectively, in BEAS-2B cells. (G and H) ROS detection kit and SOD activity assays were used to reveal the impacts between LPS treatment and let-7b-5p repression on oxidative stress in BEAS-2B cells. (I) Western blot assay was employed to determine the influences of let-7b-5p inhibitor on the protein levels of pro-c3, cleaved-c3, SOD1, SOD2 and SOD3 in LPS-induced BEAS-2B cells. (J and K) ELISA kits were purchased to disclose the influences of anti-let-7b-5p on the levels of IL-6 and IL-8 under LPS treatment in BEAS-2B cells. (L) Western blot was performed to detect the impacts between LPS and let-7b-5p inhibitor on the levels of IC3-II/IC3-I, ATG5 and P62 in BEAS-2B cells. *P<0.05, **P<0.01 and ***P<0.001.
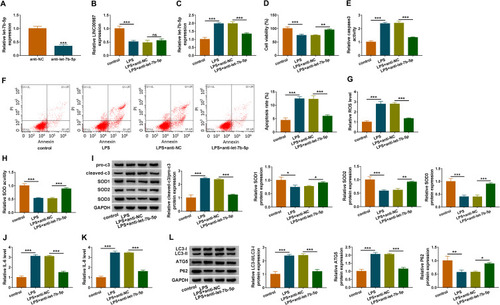
Figure 4 LINC00987 overexpression inhibited cell apoptosis, oxidative stress, inflammation and autophagy by binding to let-7b-5p in LPS-mediated BEAS-2B cells. (A) The expression levels of miRNAs possessing the binding sites of LINC00987 were determined by RT-qPCR in BEAS-2B cells transfected with LINC00987 or vector, and the binding sites between LINC00987 and let-7b-5p were predicted by lncBase online database. (B) The transfection efficiency of let-7b-5p mimic was determined by RT-qPCR. (C) Dual-luciferase reporter assay was performed to detect luciferase activity in LPS-induced BEAS-2B cells. (D) The effects between LINC00987 and let-7b-5p on let-7b-5p expression were revealed by RT-qPCR. (E) CCK-8 assay was employed to illustrate the impacts between LINC00987 and let-7b-5p on cell viability in LPS-induced BEAS-2B cells. (F and G) Caspase3 activity and apoptosis analysis assays were carried out to demonstrate the impacts between LINC00987 and let-7b-5p mimic on the apoptosis of LPS-induced BEAS-2B cells. (H and I) ROS detection kit and SOD activity assays were conducted to present the influences between LINC00987 overexpression and let-7b-5p mimic on oxidative stress in LPS-induced BEAS-2B cells. (J) The influences between LINC00987 and let-7b-5p on the expression of apoptosis-related proteins (pro-caspase3 and cleaved-caspase3) and ROS-associated proteins (SOD1, SOD2 and SOD3) were revealed by Western blot. (K and L) ELISA assays were used to disclose the impacts between LINC00987 overexpression and let-7b-5p mimic on inflammation response in LPS-induced BEAS-2B cells. (M) Western blot was performed to detect the influences between LINC00987 and let-7b-5p on the levels of LC3-II/IC3-I, ATG5 and P62 in LPS-mediated BEAS-2B cells. *P<0.05, **P<0.01 and ***P<0.001.
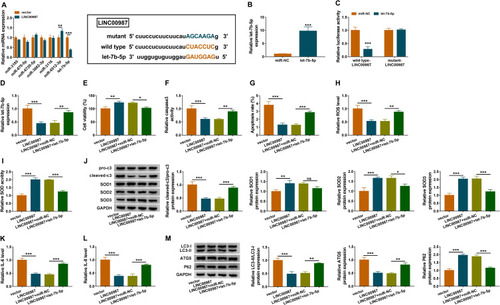
Figure 5 LINC00987 upregulated SIRT1 expression via binding to let-7b-5p. (A) MiRWalk online database was used to predict the binding sites between let-7b-5p and SIRT1 3ʹUTR. (B) Luciferase activities were detected by dual-luciferase reporter assay. (C) The effects of let-7b-5p mimic and inhibitor on SIRT1 mRNA level were revealed by RT-qPCR. (D) The effects of let-7b-5p mimic and inhibitor on SIRT1 protein level were investigated by Western blot. (E and F) The effects between LINC00987 and let-7b-5p on SIRT1 expression at mRNA and protein levels were illustrated via RT-qPCR and Western blot, respectively. ***P<0.001.
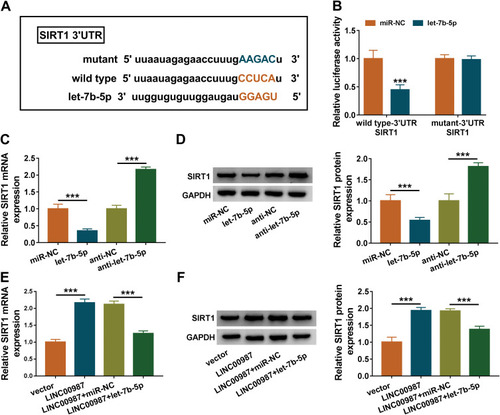
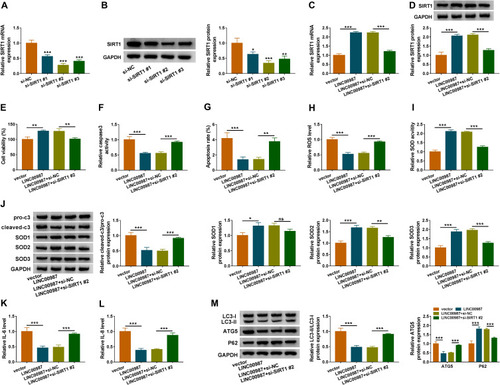
Figure 6 LINC00987 ameliorated LPS-induced disorders by regulating SITR1. (A and B) The knockdown efficiency of si-SIRT1#1, si-SIRT1#2 and si-SIRT1#3 was determined by detecting the mRNA and protein levels of SIRT1 via RT-qPCR and Western blot, respectively. (C) The effects of LINC00987 and SIRT1 knockdown on the mRNA level of SIRT1 were illustrated by RT-qPCR. (D) The effects of LINC00987 and SIRT1 knockdown on the protein expression of SIRT1 were illustrated by Western blot. (E) CCK-8 assay was performed to illustrate the effects between LINC00987 and SIRT1 silencing on cell viability in LPS-induced BEAS-2B cells. (F and G) Caspase3 activity and apoptosis analysis assays were employed to demonstrate the impacts between LINC00987 and SIRT1 deletion on cell apoptosis in LPS-induced BEAS-2B cells. (H and I) ROS detection kit and SOD activity assays were carried out to display the influences between LINC00987 overexpression and SIRT1 absence on oxidative stress in LPS-induced BEAS-2B cells. (J) The impacts between ectopic LINC00987 expression and SIRT1 downregulation on the protein expression of pro-c3, cleaved-c3, SOD1, SOD2 and SOD3 were presented by Western blot assay. (K and L) ELISA kit assays were conducted to investigate the impacts between LINC00987 overexpression and SIRT1 deletion on IL-6 and IL-8 production in LPS-induced BEAS-2B cells. (M) The effects between LINC00987 overexpression and SIRT1 knockdown on the levels of LC3-II/IC3-I, ATG5 and P62 were presented by Western blot in LPS-induced BEAS-2B cells. *P<0.05, **P<0.01 and ***P<0.001.