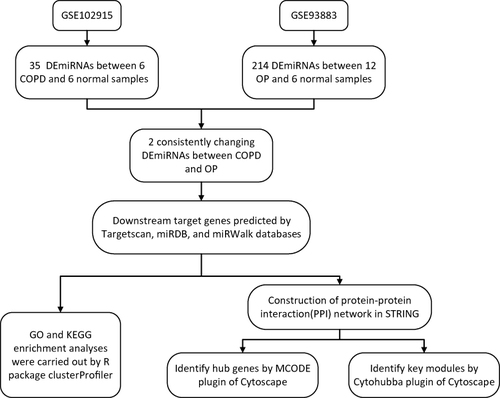
Figures & data
Table 1 Demographic and Baseline Characteristics of the Samples in the MicroRNA Array
Figure 2 Heatmaps and volcano plots of differentially expressed miRNA (|log2 fold change|>1, adjusted p-value<0.05). (A) Volcano plot of the GSE102915 dataset. (B) Volcano plot of the GSE93883 dataset. (C) Heatmap of the GSE102915 dataset. (D) Heatmap of the GSE93883 dataset. Red represents upregulated expression, blue means downregulated, and grey indicates no significant changes in volcano plot. Orange and blue in heatmaps represent patients and health control samples, respectively.
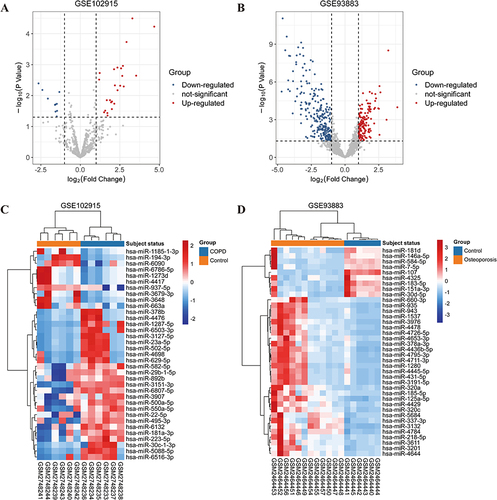
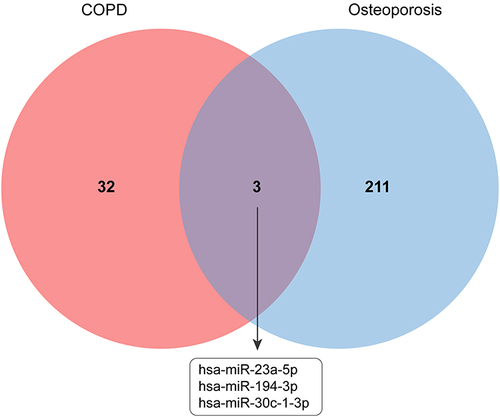
Table 2 Basic Information of Differentially Expressed miRNAs
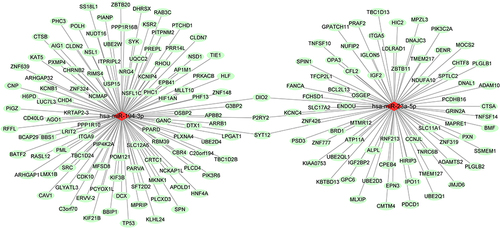
Figure 4 UpSet plot of target genes prediction.
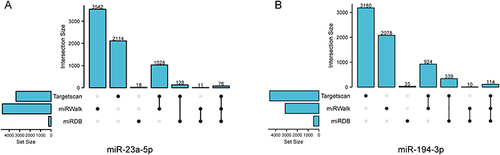
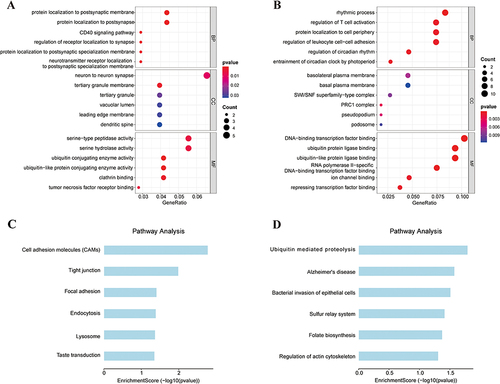
Figure 6 Results of GO term and KEGG pathways enrichment.