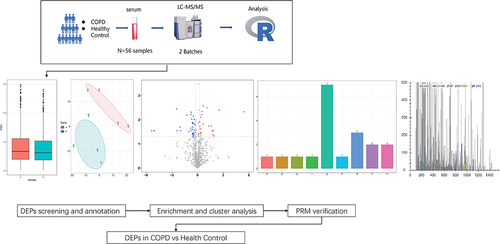
Figures & data
Table 1 Clinical Diagnoses and Demographic Characteristics of Participants
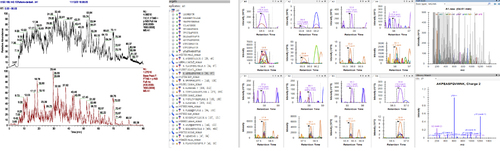
Figure 2 Process and quality control of DIA.
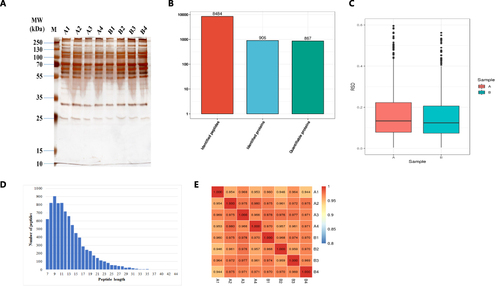
Figure 3 Results of differential expression proteins (DEPs) analysis.
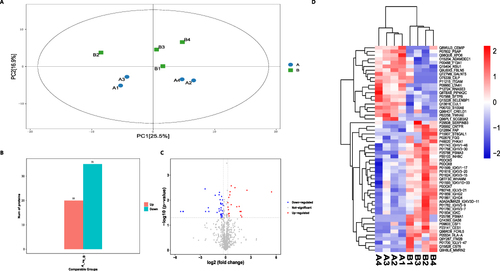
Figure 4 Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway functional enrichment of DEPs.
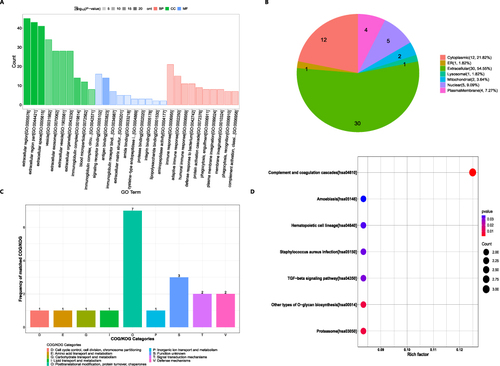
Figure 5 Protein–protein interaction (PPI) networks analysis of DEPs.
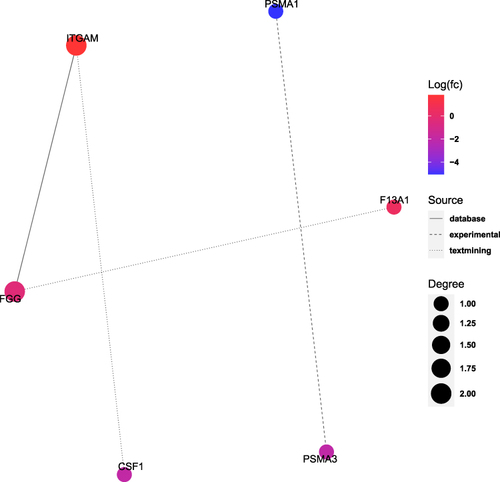
Table 2 Differential Expression Proteins in Serum Samples from COPD and Healthy Control Group