Figures & data
Figure 1 Structures of irreversible TK inhibitor.Citation15,Citation20,Citation28–Citation30 The colored ellipsoid marks the common features.
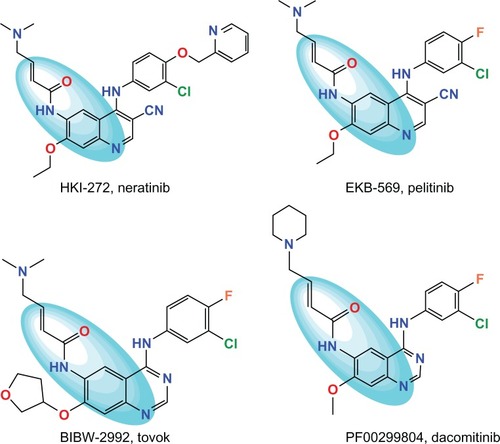
Figure 2 Schematic diagram of workflow for identifying potential EGFR-T790M inhibitors with novel scaffolds.
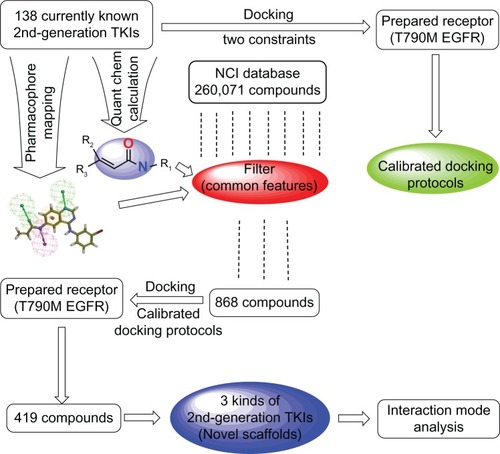
Figure 3 The common pharmacophore feature generated from compound T-001Citation37 (A) and 138 tyrosine kinase inhibitors (TKIs)Citation34–Citation37 superimposed on the feature (B). Aromatic hydrophobic center is represented by the orange wire mesh ball. Hydrogen bond accepter is depicted with the green arrow and wire mesh balls while the hydrogen bond donor is described by the pink arrow and wire mesh balls.
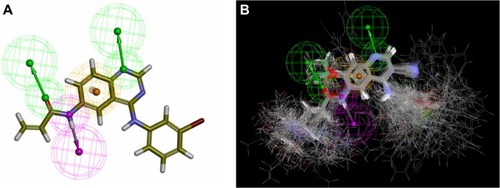
Table 1 Scoring functions employed for identifying optimal docking protocols at different stagesTable Footnote*
Table 2 NBO charge distribution and the relevant change of charge distribution in the N-electron and (N + 1)-electron molecule for N electrons systems T-005 and T-008
Figure 4 The electrophilic reactive site of some known irreversible TKIsCitation34–Citation37 calculated at B3LYP/6-31G(d) level with DFT method by employing Gaussian 03Citation40 and the effect of replacement on the bioactivity of irreversible TKI. Electrophilic center was highlighted by the red color and the nucleophilic center was painted with a blue color. Red arrow points to the β-carbon of the acrylamide group or propargyl amide group.
Abbreviations: DFT, density functional theory; TKI, tyrosine kinase inhibitor.
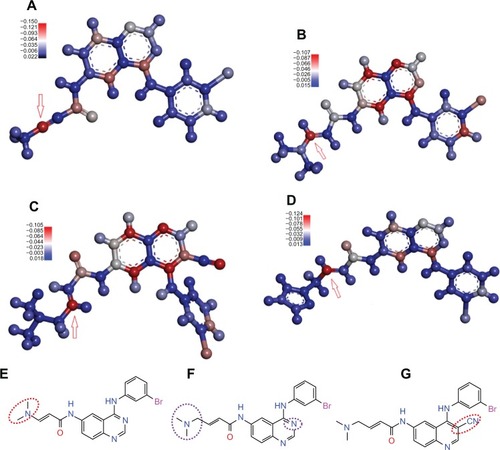
Table 3 The IC50 value and ShapegaussCitation46 score of five potent TKIsCitation34–Citation37 enriched within the top six of 138 second-generation TKIs
Table 4 The ShapegaussCitation46 score and antitumor activities of the 28 potential irreversible TKIs plus four reference irreversible TKIsCitation4,Citation37
Figure 5 In total, 21 irreversible TKIsCitation4,Citation34–Citation37 in the testing set (refer to Table S2 in supplementary materials) were mapped on the common pharmacophore feature generated from compound T-001.Citation37 T-001 is depicted by the stick and its carbon atoms are colored in green. WZ-3146Citation4 is depicted by the stick and its carbon atoms are colored pink; WZ-4002Citation4 is depicted by the stick and its carbon atoms are colored sky blue; WZ-8040Citation4 is depicted by the stick and its carbon atoms are colored blue.
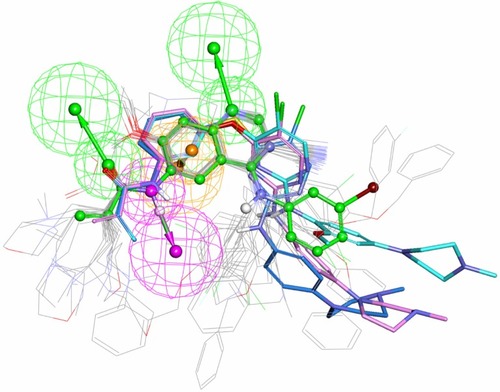
Figure 6 Potential irreversible TKIs that had been identified from the NCI database (refer to ) were mapped onto the common pharmacophore feature generated from reference compound T-001.Citation37 T-001 is depicted by the stick and its carbon atoms are colored green. Representative compounds NSC-621158, NSC-642576, and NSC-57443 are depicted by the stick and their carbon atoms are colored pink, sky blue, and blue, respectively.
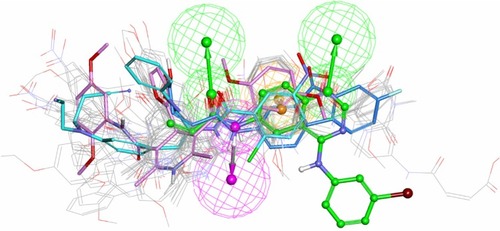
Figure 7 Three classes of potential irreversible TKIs with novel scaffolds and their corresponding representative molecules.
Abbreviations: NSC, Nomenclature Standards Committee; TKI, tyrosine kinase inhibitor.
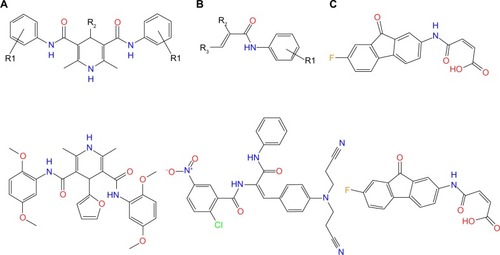
Figure 8 Pharmacophore modelCitation27,Citation51 of the ATP binding site in tyrosine kinase cSrc (PDB ID: 2QQ7).Citation44 This site is represented by the solid surface. Adenine region, sugar region, hydrophobic region I, and hydrophobic region II are colored pink, yellow, blue, and green, respectively.
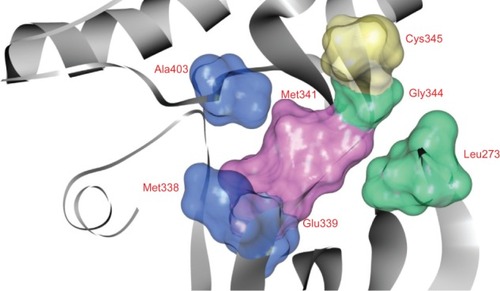
Figure 9 The illustrations showing the interaction between the model system and three representative compounds as well as between the reference compound T-001Citation37 and the model system. Residues are displayed in thin stick style. Carbon atoms of residues corresponding to adenine region, sugar region, hydrophobic region I, and hydrophobic region II are colored pink, yellow, blue, and light green, respectively. Compounds are represented by a thick stick and their carbon atoms are colored green. Interactions such as hydrogen bond, cation-π interaction, and π-σ interaction are highlighted.
Abbreviations: Ala, alanine; Gln, glutamine; Glu, glutamic acid; Met, methionine; Thr, threonine; Leu, leucine; Gly, glycine; Cys, cysteine.
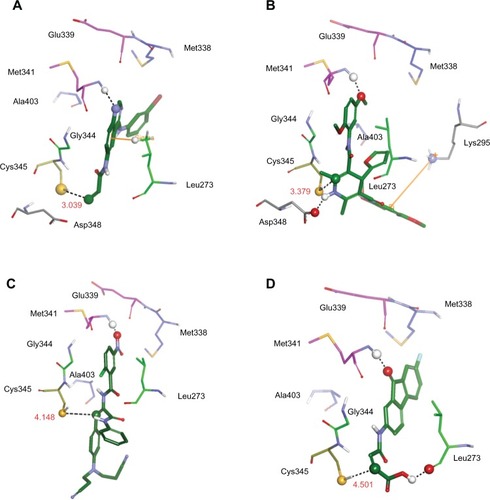
Table S3 The structures of 28 potential irreversible TKIs with three classes of novel scaffolds identified by docking molecules from NCI database (release 3 files) to the binding site of T790M EGFR using FRED 2.2.5Citation1 (OpenEye Scientific Software Inc., Santa Fe, NM, USA).