Figures & data
Figure 1 Schematic on GLP-1 secretion in the L-cell. Glucose arising from carbohydrate metabolism is transported at the luminal face of the L-cell via sodium-glucose cotransporter-1 (SGLT-1), which is coupled with Na+ influx, depolarizing the cell membrane (ΔΨ), opening VDCC, increasing intracellular Ca2+ levels, and triggering exocytosis of GLP-1 containing granules at the L-cell’s basolateral face. Increased intracellular ATP from glucose metabolism closes KATP channels, potentiating GLP-1 release. Long-chain fatty acids interact with G protein-coupled receptors (GPR40 and GPR120), triggering intracellular Ca2+ release to prompt GLP-1 release. Amino acids affect the secretion of GLP-1 by a similar mechanism mediated by Na+ ions.
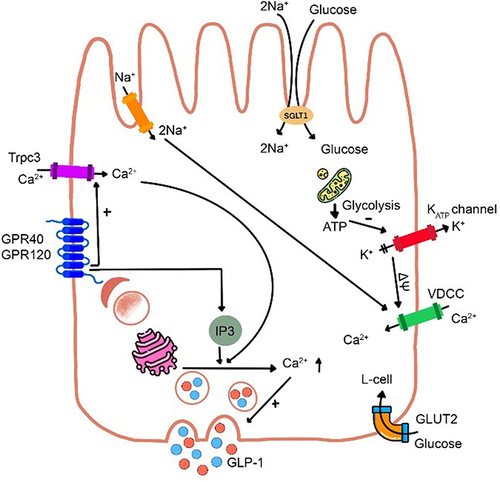
Figure 2 (a) 3D structure of human dipeptidyl peptidase-IV in complex with a potent selective inhibitor, alogliptin (indicated by red arrow) (PDB ID 2ONC; PDB DOI https://doi.org/10.2210/pdb2ONC/pdb; Resolution 2.55 Å; deposited by Feng et al, 2007. Total structure weight: 346.99 kDa); (b) DPP-IV binding subsites for the inhibitors which are numbered from the cleavage point to the S2, S1, S1’, S2’, and S2 extensive subsites. Class I inhibitors bind to S1 and S2 subsites; Class II inhibitors to S2’, S1’, and S2 subsites; Class III inhibitors to S1, S2, and S2 extensive subsites. Adapted from Biochem Biophys Res Commun, 434(2), Nabeno M, Akahoshi F, Kishida H, et al. A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site. 191–196, Copyright (2013), with permission from Elsevier.Citation35
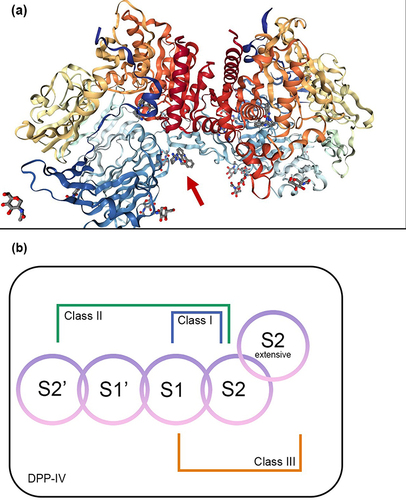
Table 1 In silico Study
Table 2 In vitro Study Using Human DPP-IV Inhibitory Screening Kit
Table 3 In vivo Study
Table 4 Studies in Human
