Figures & data
Figure 1 Vpr expression kinetics and generation of BiFC in transfected cells.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; VN-Vpr, Vpr fused to N-terminus of Venus protein; VC-Vpr, Vpr fused to C-terminus of Venus protein; HA, HA-tagged Vpr.
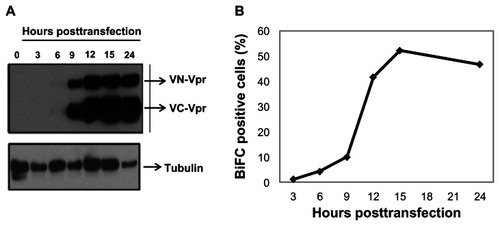
Figure 2 Competition assay to detect the loss of BiFC signal generated by Vpr dimerization.
Abbreviations: BiFC, bimolecular fluorescence complementation; Vpr, viral protein R; DNA, deoxyribonucleic acid.
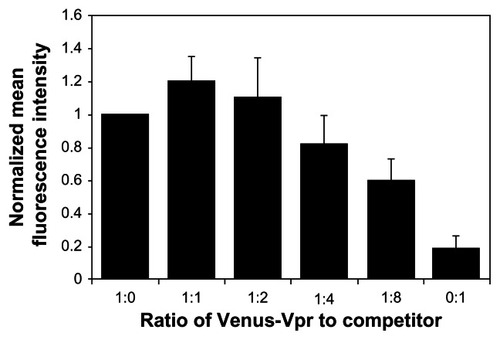
Figure 3 Fluorescence intensity of BiFC generated by Venus-Vpr in the presence of competitor untagged Vpr.
Abbreviations: BiFC, bimolecular fluorescence complementation; Vpr, viral protein R; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride.
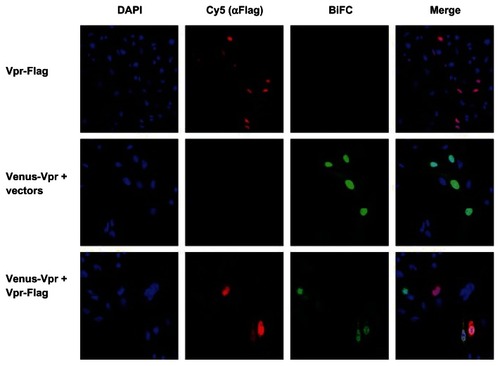
Figure 4 Ability of Vpr peptides to interfere with Vpr dimerization.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; PBS, phosphate buffered saline; DDDT, dimethyl sulfoxide; TF, transfection.
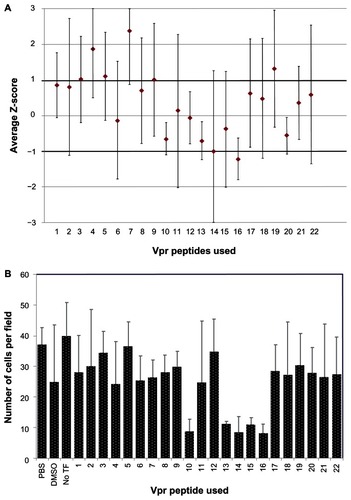
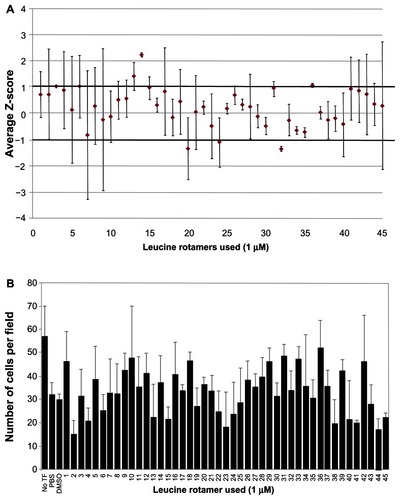
Figure 5 Ability of leucine rotamer library to inhibit Vpr dimerization measured by BiFC signal.
Abbreviations: Vpr, viral protein R; BiFC, bimolecular fluorescence complementation; TF, transfection; PBS, phosphate buffered saline; DDDT, dimethyl sulfoxide.