Figures & data
Figure 1 The miRNA biogenesis pathway.
Abbreviations: Ago, argonaute; GTP, guanosine triphosphate; miRISC, miRNA-induced silencing complex; miRNA, microRNA; pri-miRNA, primary miRNA; pre-miRNA, precursor miRNA; TRBP, trans-activation response element RNA binding protein.
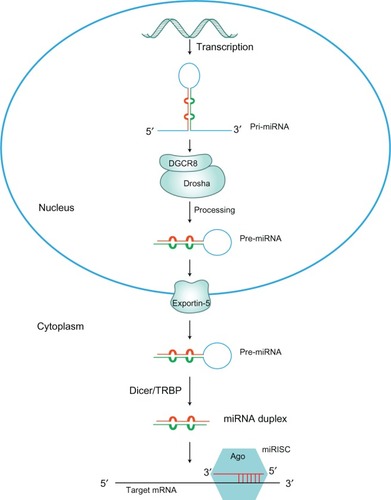
Figure 2 Putative mechanisms of α-synuclein neurodegeneration in familial PD.
Abbreviations: PD, Parkinson’s disease; UCH-L1, ubiquitin carboxy-terminal hydrolase L1; UPP, ubiquitin proteasome pathway.
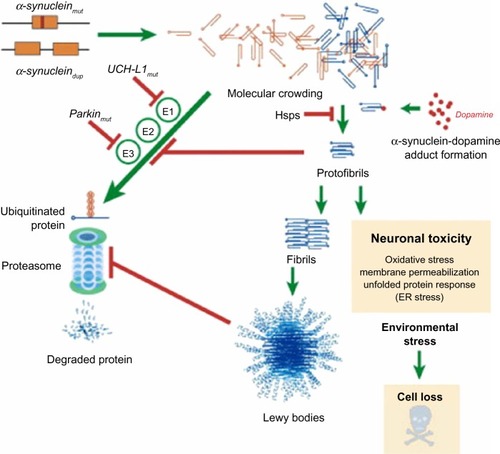
Figure 3 MicroRNA (miR-7 and miR-153) regulation of SNCA in PD.
Abbreviations: 3′-UTR, 3′ untranslated region; α-syn, α-synuclein; mRNA, messenger RNA; miRNA, microRNA; PD, Parkinson’s disease.
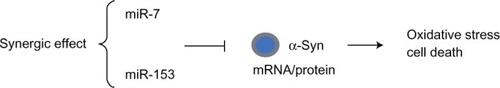
Figure 4 LRRK2 regulates the microRNA (let-7 and miR-184*) network.
Abbreviation: LRRK2, leucine-rich repeat kinase 2.
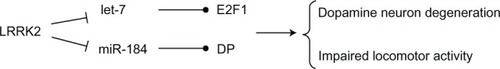
Figure 5 The role of PARK2 and PARK7 in the pathogenesis of PD.
Abbreviation: PD, Parkinson’s disease.
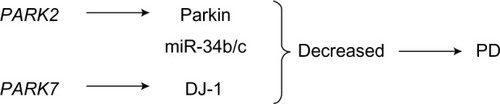
Figure 6 miR-133b and Pitx3 form a negative feedback loop.
Abbreviations: DA, dopaminergic; Pitx3, pituitary homeobox 3.
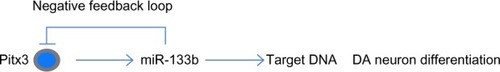
Figure 7 MicroRNA (miR-433) regulation of FGF20 in PD.
Abbreviations: α-syn, α-synuclein; FGF20, fibroblast growth factor 20; PD, Parkinson’s disease; SNP, single-nucleotide polymorphism.

Table 1 miRNAs potentially involved in PD pathophysiology
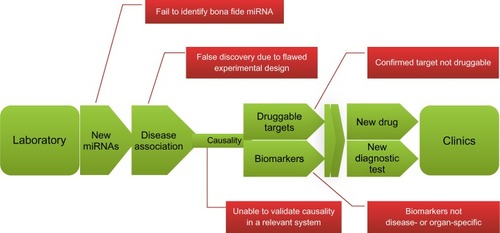
Figure 8 The road from laboratory to clinic: the promises and challenges of miRNA research.
Abbreviation: miRNA, microRNA.