Figures & data
Figure 1 Work flow of present study.
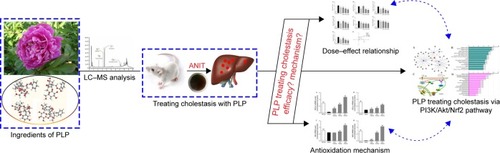
Table 1 Primers sequences for RT-PCR
Table 2 Identification of compounds present in PLP by UHPLC-Q-TOF
Figure 2 Positive ES mode of LC–MS of PLP aqueous extract.
Abbreviations: ES, electron spray; PLP, Paeonia lactiflora Pall.; LC–MS, liquid chromatography–mass spectrometry; tR, retention time.
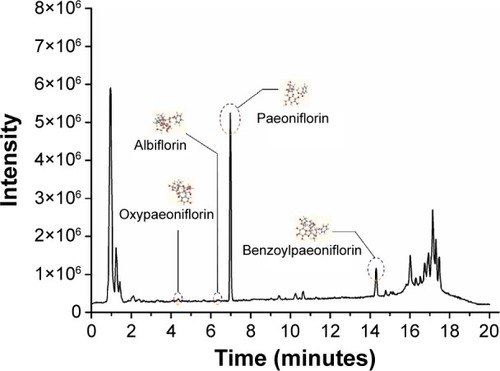
Figure 3 Effects of PLP on serum indices of liver function.
Abbreviations: PLP, Paeonia lactiflora Pall.; ANIT, alpha-naphthylisothiocyanate; UDCA, ursodeoxycholic acid; ALT, alanine aminotransferase; AST, aspartate aminotransferase; TBIL, total bilirubin; DBIL, direct bilirubin; TBA, total bile acid; ALP, alkaline phosphatase; γ-GT, γ-glutamyl transpeptidase; PCA, principal component analysis; SD, standard deviation.
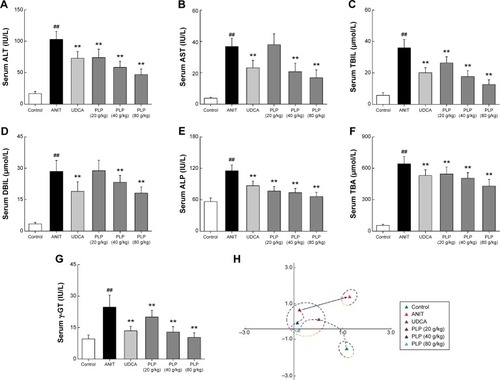
Figure 4 Effect of PLP on histological changes in the liver tissue of ANIT-induced rats.
Abbreviations: PLP, Paeonia lactiflora Pall.; ANIT, alpha-naphthylisothiocyanate; UDCA, ursodeoxycholic acid; HE, hematoxylin-eosin.
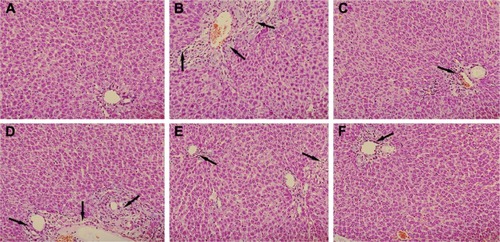
Figure 5 GSH level and Western blot analysis of pAkt, GCLc, GCLm, and Nrf2.
Abbreviations: PLP, Paeonia lactiflora Pall.; ANIT, alpha-naphthylisothiocyanate; UDCA, ursodeoxycholic acid; Nrf2, NF-E2-related factor 2; GSH, glutathione; GCLc, glutamate-cysteine ligase catalytic subunit; GCLm, glutamate-cysteine ligase modulatory subunit; SD, standard deviation.
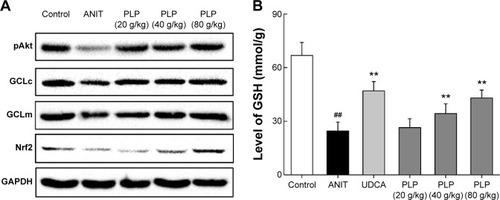
Figure 6 mRNA expessions of GLCc, GLCm, Nqo1, HO-1, Akt, and Nrf2 (%).
Abbreviations: PLP, Paeonia lactiflora Pall.; ANIT, alpha-naphthylisothiocyanate; UDCA, ursodeoxycholic acid; Nrf2, NF-E2-related factor 2; GCLc, glutamate-cysteine ligase catalytic subunit; GCLm, glutamate-cysteine ligase modulatory subunit; HO-1, heme oxygenase-1; NQO1, NAD(P)H/quinone oxidoreductase 1; SD, standard deviation.
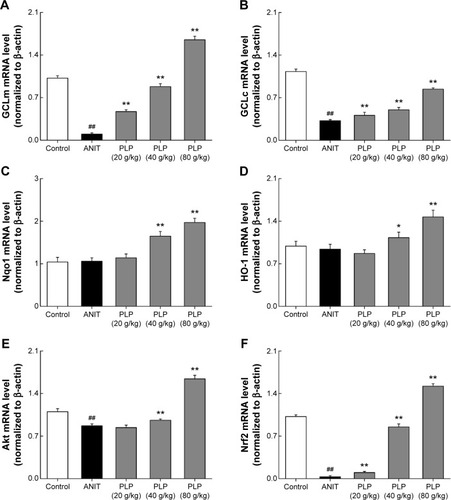
Figure 7 PLP putative targets network validation and its possible mechanism.
Abbreviations: PLP, Paeonia lactiflora Pall.; ANIT, alpha-naphthylisothiocyanate; Nrf2, NF-E2-related factor 2; GSH, Glutathione; GCLc, glutamate-cysteine ligase catalytic subunit; GCLm, glutamate-cysteine ligase modulatory subunit.
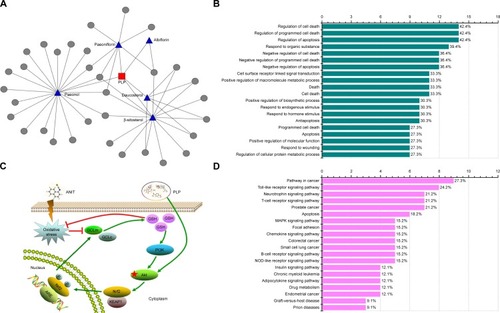
Figure S1 Mass-to-charge (m/z) information of four compounds.
Table S1 PLP putative targets
