Figures & data
Table 1 The information of primer sequences
Figure 1 Expression of MALAT1 in cardiomyocytes after treatment with palmitic acid (PA). *P<0.05. Data are presented as means ± standard error from three independent experiments.
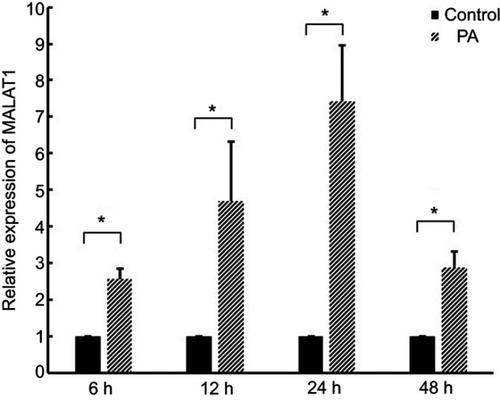
Figure 2 Effect of the knockdown of MALAT1 on palmitic acid (PA)-induced myocardial injury. (A) The expression of MALAT1 in AC16 cells was detected by RT-qPCR after transfection with MALAT1-specific siRNA (si-MALAT1) and a negative control (scramble). (B) Cell viability was determined by a Cell Counting Kit-8 assay. (C) Lactate dehydrogenase (LDH) activity in the culture medium was detected by spectrophotometry. (D) Creatine kinase-MB (CK-MB) activity in the culture medium was detected by spectrophotometry. (E) Apoptosis was measured by flow cytometry. (F) Statistical analysis of the ratio of apoptotic cells. *P<0.05. Data are presented as the mean ± standard error from three independent experiments.
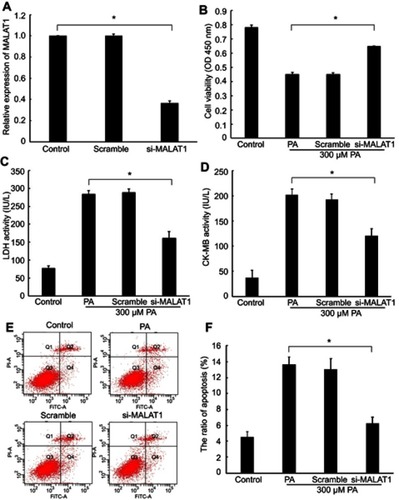
Figure 3 Interaction between MALAT1 and miR-26a. (A) miRNA binding sites in the MALAT1 sequence. (B) Luciferase reporter gene assay validated the interaction between MALAT1 and miR-26a in AC16 cells. pmirGLO-MALAT1, luciferase vector of MALAT1; pmirGLO-MALAT1-mut, MALAT1 mutant vector. (C) Effect of MALAT1 on the level of miR-26a. (D) Effect of miR-26a on the level of MALAT1. *P<0.05. Data are presented as the mean ± standard error from three independent experiments.
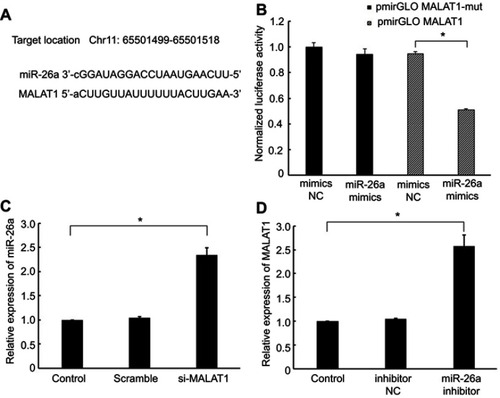
Figure 4 The regulatory effect of MALAT1 on the HMGB1/TLR4/NF-κB signaling pathway was mediated by miR-26a. MALAT1 negative control (scramble) and MALAT1-specific siRNA (si-MALAT1) alone or in combination with an miR-26a inhibitor or miR-26 negative control (inhibitor NC) was transfected into AC16 cells, followed by palmitic acid treatment. (A) The mRNA expression of HMGB1 was detected by RT-qPCR. (B) The protein expression of HMGB1 was detected by western blotting. (C) The protein expression of TLR4 was detected by western blotting. (D) The protein expression of NF-κB (P65) in the nucleus was detected by western blotting. (E) The protein expression of NF-κB (P65) in the cytoplasm was detected by western blotting. (F) The activity of NF-κB was detected by ELISA. *P<0.05. Data are presented as the mean ± standard error from three independent experiments.
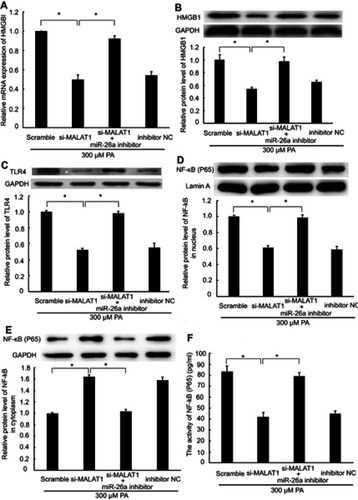
Figure 5 The effect of MALAT1 on PA-induced myocardial inflammation was mediated by miR-26a. MALAT1 negative control (scramble) and MALAT1-specific siRNA (si-MALAT1) alone or in combination with an miR-26a inhibitor or miR-26 negative control (inhibitor NC) and TNF-α-specific siRNA (si-TNF-ɑ) were transfected into AC16 cells, followed by palmitic acid (PA) treatment. (A) The mRNA expression of TNF-α was detected by RT-qPCR. (B) The mRNA expression of IL-1β was detected by RT-qPCR. (C) The concentration of TNF-α in the culture medium was detected by ELISA. (D) The concentration of IL-1β in the culture medium was detected by ELISA. *P<0.05. Data are presented as the mean ± standard error from three independent experiments.
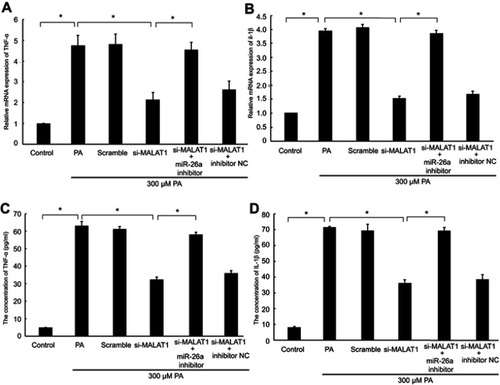
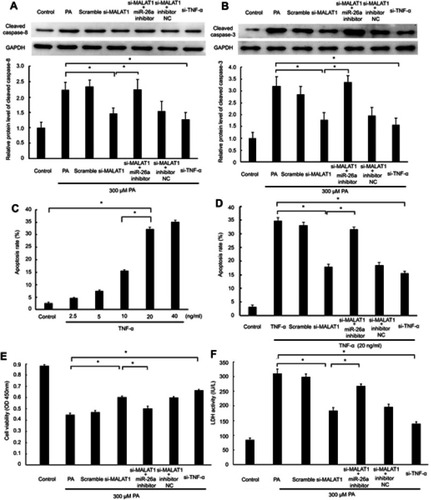
Figure 6 MALAT1/miR-26a manipulation regulated PA-induced cell death via a TNF-α-induced apoptosis pathway. MALAT1 negative control (scramble) and MALAT1-specific siRNA (si-MALAT1) alone or in combination with an miR-26a inhibitor or miR-26 negative control (inhibitor NC) and TNF-α-specific siRNA (si-TNF-ɑ) were transfected in AC16 cells, followed by palmitic acid (PA)/TNF-α treatment. (A) The protein expression of cleaved caspase-8 was detected by western blotting. (B) The protein expression of cleaved caspase-3 was detected by western blotting. (C) AC16 cells were treated with different concentrations of TNF-α for 12 h to induce apoptosis. (D) MALAT1/miR-26a manipulation regulated TNF-α-induced apoptosis. (E) Cell viability was determined by CCK8 assay. (F) Lactate dehydrogenase (LDH) activity in the culture medium was detected by spectrophotometry. *P<0.05. Data are presented as the mean ± standard error from three independent experiments.