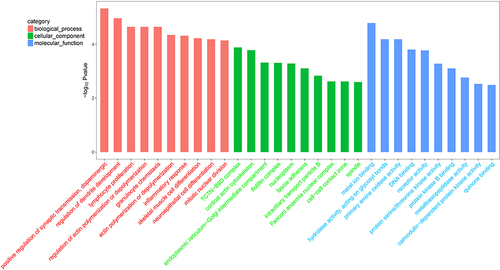
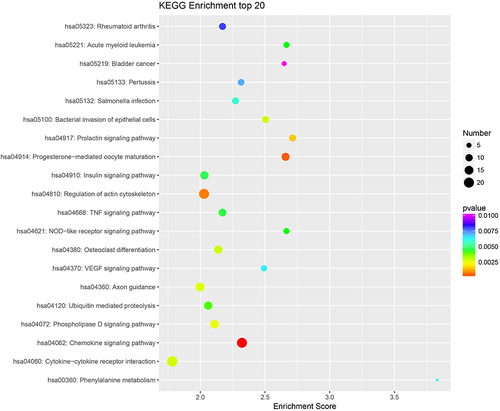
Figures & data
Table 1 Primer Sequences
Figure 1 The volcano diagram of differential expression.
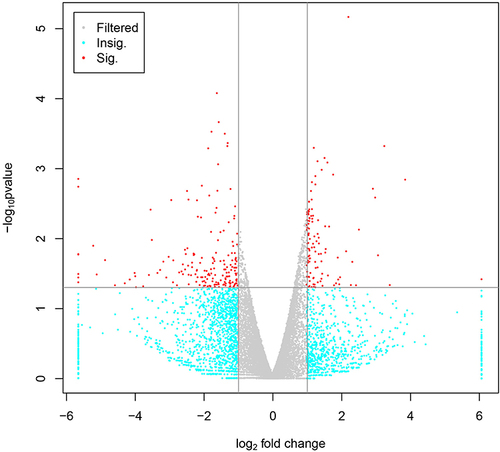
Figure 2 The cluster diagram of differential expression.
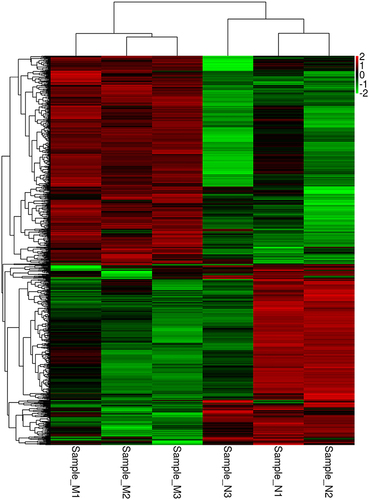
Table 2 The Results of Screening of the Differentially Expressed Genes
Table 3 The mRNA Expression Analysis in the Validation of Candidate Genes (, n=20)
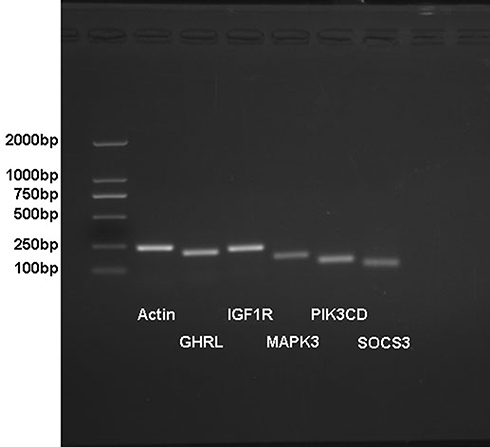
Figure 5 The electropherogram of PCR primer identification.