Figures & data
Table 1 Primer Sequences Used for PCR
Figure 1 Circ_Arf3 down-regulation in MCs treated with HG. (A) qRT-PCR was employed to examine the expression of circ_Arf3 in MCs treated with HG or normal control. (B) As shown in nuclear RNA fractionation and cytoplasmic experiments, circ_Arf3 was mainly found in the cytoplasm of HG-treated MCs. (C) RNase R degradation was used to detect the stability of circ_Arf3. (D) Actinomycin D treatment assay was used to detect the stability of circ_Arf3. *P<0.05.
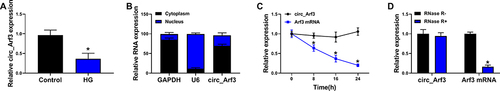
Figure 2 Circ_Arf3 overexpression reduced HG-induced proliferation and increase of fibrosis-related proteins in mouse MCs. (A) The transfection efficiency of circ_Arf3 overexpression plasmid or control vector was investigated by qRT-PCR assay. (B–E) MCs were transfected with or without circ_Arf3 overexpression plasmid or control vector and then treated with HG or normal control. (B) EDU proliferation assay was performed to determine cells proliferation. (C) CCK-8 was used to detect cell viability. (D) The expression level of PCNA was determined by Western blot. (E) The expressions of DN fibrosis-related proteins α-SMA, Col I, FN and Col IV were detected by Western blot. *P<0.05.
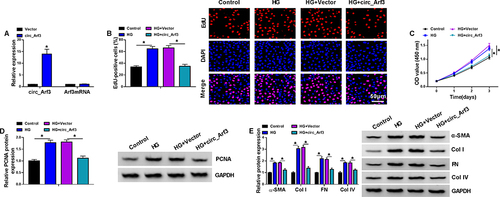
Figure 3 Circ_Arf3 targeted regulating miR-452-5p. (A) The complementary sequences between miR-452-5p and circ_Arf3 were shown. (B) Dual-luciferase reporter assays were performed to confirm the association between miR-452-5p and circ_Arf3. (C) The relationship between miR-452-5p and circ_Arf3 was verified by RNA pull-down assay. (D) qRT-PCR was carried out to measure the effect of circ_Arf3 on miR-452-5p expression. (E) qRT-PCR was carried out to measure the effect of HG treatment on miR-452-5p expression. *P<0.05.
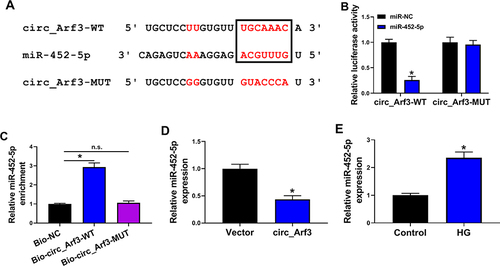
Figure 4 MiR-452-5p reversed the effect of circ_Arf3 up-regulation. (A) The expression of miR-452-5p was examined using qRT-PCR assay in MCs transfected with circ_Arf3 overexpression plasmid, control vector, circ_Arf3 overexpression plasmid+miR-NC mimic or circ_Arf3 overexpression plasmid+miR-452-5p mimic. (B–E) MCs were transfected with or without circ_Arf3 overexpression plasmid, control vector, circ_Arf3 overexpression plasmid+miR-NC mimic or circ_Arf3 overexpression plasmid+miR-452-5p mimic and then treated with HG or normal control. (B) EDU proliferation assay was performed to determine cell proliferation. (C) CCK-8 was used to detect cell viability. (D) The expression level of PCNA was determined by Western blot. (E) The expressions of DN fibrosis-related proteins α-SMA, Col I, FN and Col IV were detected by Western blot. *P<0.05.
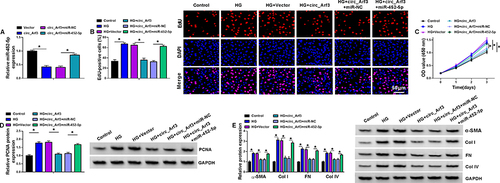
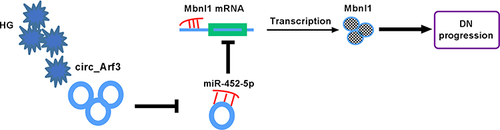
Figure 5 MiR-452-5p targeted Mbnl1. (A) The complementary sequences between miR-452-5p and Mbnl1 were shown. (B) Dual-luciferase reporter assays were performed to confirm the association between miR-452-5p and Mbnl1. (C) The relationship between miR-452-5p and Mbnl1 was verified by RNA pull-down assay. (D) qRT-PCR was carried out to measure the expression of miR-452-5p. (E) qRT-PCR was carried out to measure the expression level of Mbnl1 in different treatments. (F) Western blot was carried out to measure the expression level of Mbnl1 in different treatments. (G and H) The expression of Mbnl1 in HG-treated cells was determined by qRT-PCR and Western blot. (I and J) The expression of Mbnl1 was determined with transfection of circ_Arf3 or circ_Arf3 and miR-452-5p by qRT-PCR and Western blot. *P<0.05.
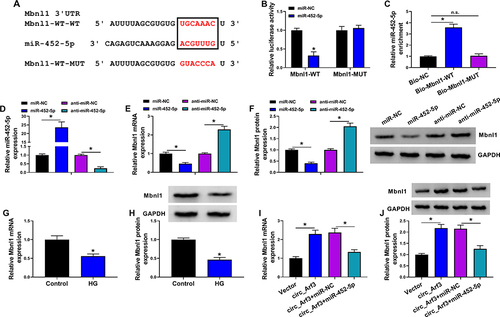
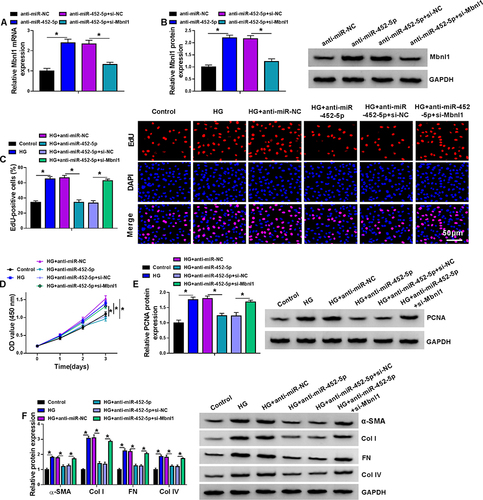
Figure 6 The miR-452-5p/Mbnl1 axis regulated HG-induced proliferation and fibrosis-related protein expression in mouse MCs. (A and B) The expression levels of Mbnl1 mRNA and protein were detected by qRT-PCR and Western blot in MCs transfected with anti-miR-NC, anti-miR-452-5p, anti-miR-452-5p+si-NC or anti-miR-452-5p+si-Mbnl1. (C–F) MCs were transfected with or without anti-miR-NC, anti-miR-452-5p, anti-miR-452-5p+si-NC or anti-miR-452-5p+si-Mbnl1 and then treated with HG or normal control.(C) EDU proliferation assay was performed to determine cell proliferation. (D) CCK-8 was used to detect cell viability. (E) The expression level of PCNA was determined by Western blot. (F) The expressions of DN fibrosis-related proteins α-SMA, Col I, FN and Col IV were detected by Western blot. *P<0.05.