Figures & data
Table 1 Clinical Characteristics Between DPN Group and T2DM Group
Table 2 The Use of Drugs in T2DM Group and DPN Group
Figure 1 Representative 600‐MHz 1H nuclear magnetic resonance spectrum (δ0.0~8.75,) of the T2DM group (blue) and DPN group (purple). All peaks were referenced to the resonance of TSP at 0 ppm. Free EDTA signals in all samples indicated that all Ca2+ and Mg2+ were chelated. Ca-EDTA, Mg-EDTA were EDTA complexes.
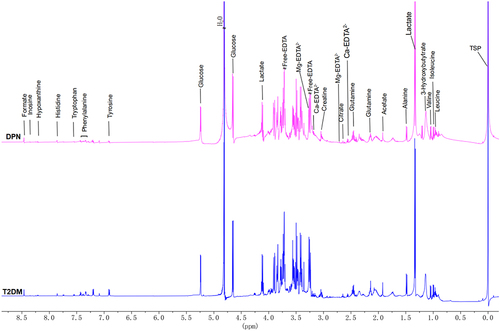
Figure 2 A heat map revealed the relative concentrations of twenty metabolites between the T2DM group and DPN group. The different colors were corresponding to different values in the right palette (0–100%).
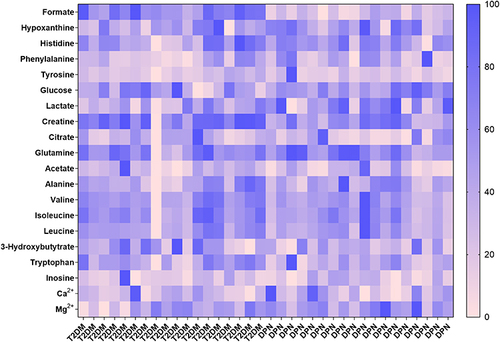
Figure 3 Principal components analysis (PCA) score plot (a) and OPLS-DA scores plot (b) indicated discrimination of the metabolites between the T2DM group and DPN group. Permutation test plot verified the OPLS-DA model (c): Q2 (cum)=0.658, R2X (cum)=0.523, R2Y (cum)=0.826. The abscissa of the model verification permutation test plot represents the accuracy of the model, the ordinate represents the frequency of the accuracy of 200 models in the 200 permutation tests, and the arrows indicate the location of the accuracy of this OPLS-DA model. R2X and R2Y respectively indicate the explanatory rate of the built model to the X and Y matrices, Q2 represents the predictive ability of the model. The closer the values of R2 and Q2 to 1, the better the model.
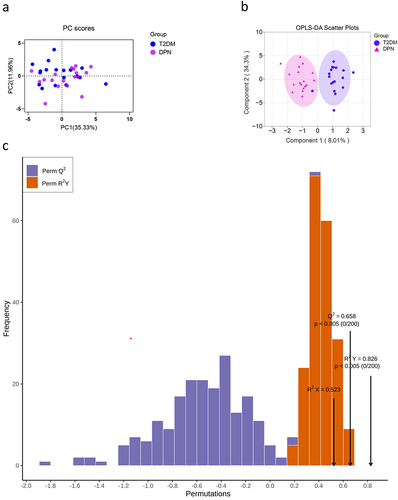
Table 3 Twenty Serum Metabolites Concentration and VIP Scores Between the T2DM Group and DPN Group
Table 4 Correlation Between Formate and Other Clinical Characteristics
Table 5 Stepwise Multiple Linear Regression Analysis of DPN and Serum Indexes
Table 6 The Coefficients and Constant of Predictive Formula Based on Stepwise Multiple Regression Analysis
Figure 4 The heat map showed diagnostic ability of each metabolite with AUC values (a). The different colors were corresponding to different AUC values in the right palette. Formate showed the highest AUC value in all the metabolites for the diagnosis of DPN (b).
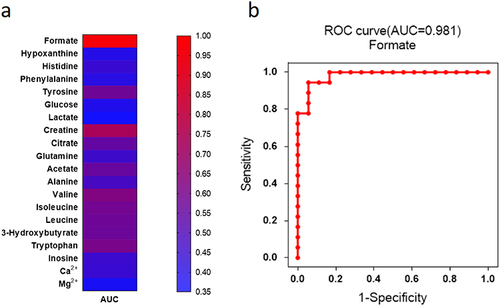
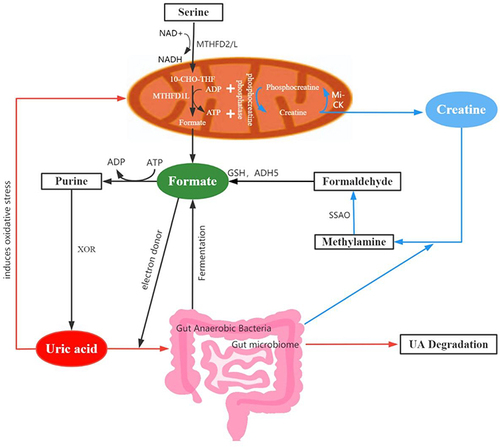
Figure 5 Potential mechanisms of formate in the development of DPN. Dysfunction of mitochondria might decrease the production of formate (black arrows) and then limit the electron supplement in uric acid metabolism (Orange arrows). Gut anaerobic bacteria (black arrows) and creatine (blue arrows) could also produce a certain proportion of formate.