Figures & data
Table 1 Epidemiological data from S. pneumoniae strains isolated from children
Figure 1 Phylogenetic relationships of S. pneumoniae isolates based on single-nucleotide polymorphisms from whole DNA sequences.
Abbreviations: PEN, penicillin; CXM, cefuroxime; CRO, ceftriaxone; MAC, macrolides (erythromycin and azithromycin) and clindamycin; SXT, sulfamethoxazole–trimethoprim.
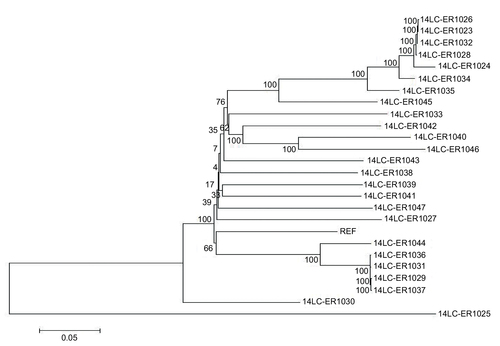
Figure 2 Principle component analysis of the whole genomes of 25 pneumococcal isolates.
Abbreviations: PEN, penicillin; CXM, cefuroxime; CRO, ceftriaxone; MAC, macrolides (erythromycin and azithromycin) and clindamycin; SXT, sulfamethoxazole–trimethoprim.
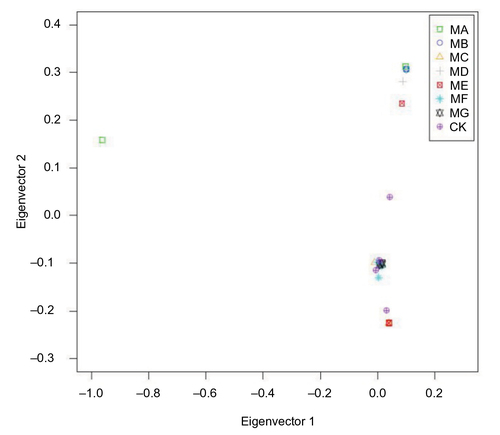
Table 2 Indel changes closely associated with antibiotic resistance in S. pneumoniae
Table 3 SNP changes associated with antibiotic resistance in S. pneumoniae
Figure 3 Indel and SNP variation in the complete genome of S. pneumoniae R6 compared with 25 S. pneumoniae clinical isolates.
Abbreviation: SNP, single-nucleotide polymorphisms.
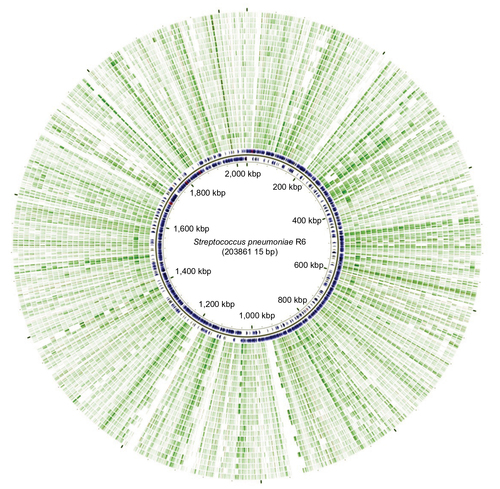
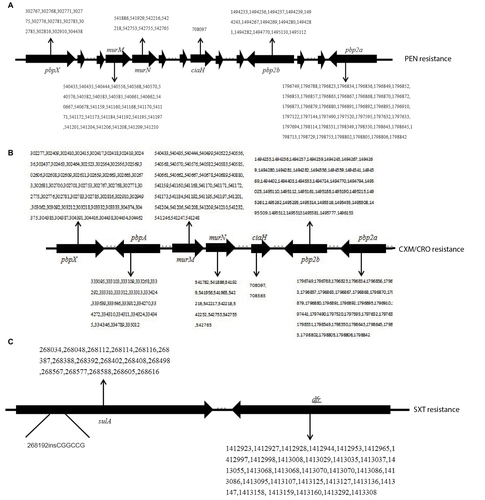
Figure 4 (A) SNPs changes in PBPs associated with penicillin resistance in S. pneumoniae. (B) SNPs changes in PBPs associated with cephalosporins resistance in S. pneumoniae. (C) SNP changes associated with sulfamethoxazole–trimethoprim resistance in S. pneumonia.