Figures & data
Figure 1 Functional domain and conservation of rpsL (A) and gyrA (B) mutation sites in Mycobacterium spp.
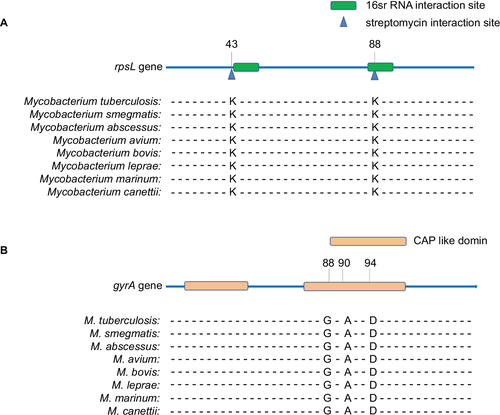
Table 1 Mutation types of single- and dual-resistant Mycobacterium smegmatis mutants under the selection of antibiotics
Figure 2 Relative fitness of Mycobacterium smegmatis mutants.
Abbreviations: FLQ, fluoroquinolones; STR, streptomycin.
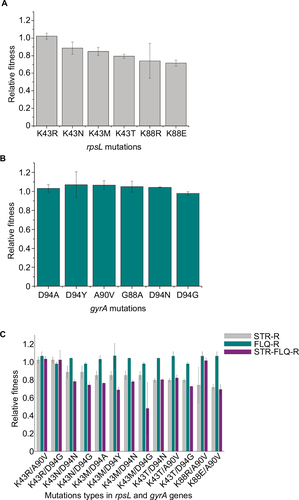
Figure 3 Frequency of dual-mutation of rpsL and gyrA found in clinical Mycobacterium tuberculosis isolates.
Abbreviation: MTB, Mycobacterium tuberculosis.
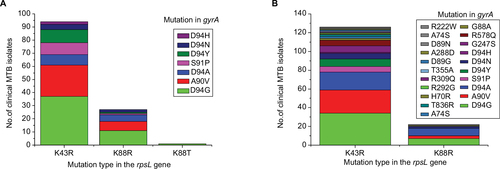
Table 2 Gene mutations in clinical STR- and FLQ-resistant Mycobacterium tuberculosis isolates
Figure S1 Model of mutations appearing in rpsL and gyrA in this study.
Table S1 Epistasis (ε) in mutants resistant to STR and FLQ

