Figures & data
Table 1 Demographic data of the 39 HIV-1-infected patients with virological failure to STRs
Figure 1 Drug resistance according to the HIVdb program of Stanford University among 39 HIV-1-infected patients with virological failure to STRs.
Abbreviations: INSTI, integrase strand transfer inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; STR, single-tablet regimen.
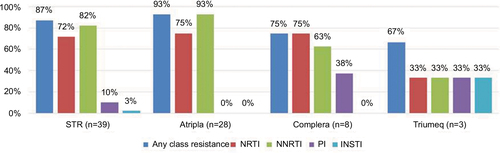
Figure 2 The prevalence of drug resistance to NRTI, NNRTI, PI, and INSTI among 39 HIV-1-infected patients with virological failure to STRs.
Abbreviations: INSTI, integrase strand transfer inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; STR, single-tablet regimen.
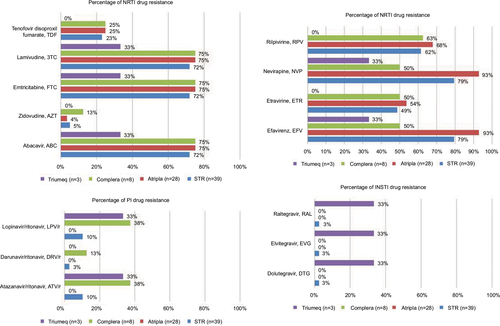
Figure 3 Percentage of HIV drug resistance-associated mutations to NRTI, NNRTI, PI, and INSTI among the 39 HIV-1-infected patients with virological failure to STRs.
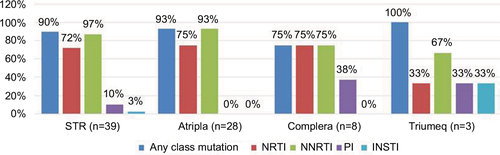
Figure 4 The prevalence of drug resistance-associated mutations to NRTI, NNRTI, PI, and INSTI among the 39 HIV-1-infected patients with virological failure to STRs.
Abbreviations: INSTI, integrase strand transfer inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; STR, single-tablet regimen.
Table 2 Clinical manifestations among five STR users with virological failure without drug resistance
Table 3 Demographic information, antiretroviral treatment duration, and resistance between 8 treatment-naïve patients initiating an STR and 31 treatment-experienced patients switching to an STR with virological failure
Table 4 Risk factors associated with TDF resistance in univariate analysis
Table 5 Risk factors associated with PI resistance among the 39 patients with STR failure
