Figures & data
Figure 1 Identification of E. coli with or without lysis buffer.
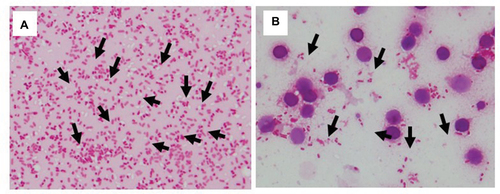
Figure 2 Display within the BioTyper 2.0 graphic view in the case of E. coli.
Abbreviation: MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
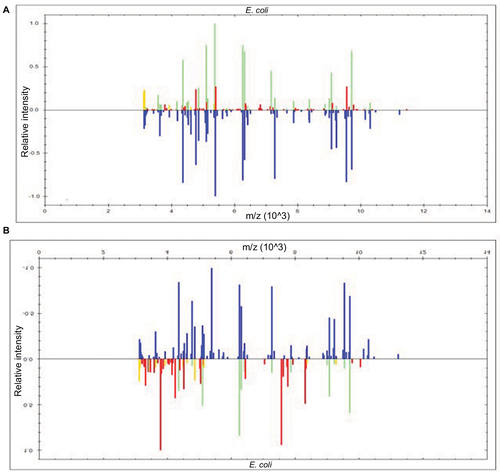
Table 1 Detection efficiency of bacteria from blood culture broth by TOF-MS with or without lysis buffer
Figure 3 Identification of S. pneumoniae with or without lysis buffer.
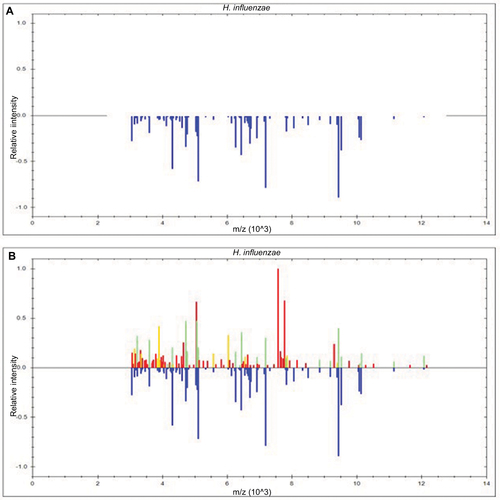
Figure 4 Identification of H. influenzae with or without lysis buffer.
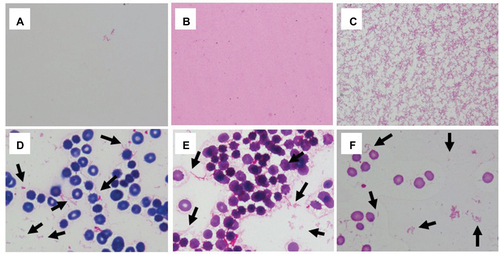
Figure 5 Display within the BioTyper 2.0 graphic view in the case of H. influenzae.
Abbreviation: MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.