Figures & data
Table 1 List of primers used in this study
Table 2 Antibiotic susceptibility testing of enterococcal clinical isolates
Figure 1 Genotyping of E. faecalis clinical isolates using RAPD-PCR method.
Notes: Dendrogram was created using RAPD-PCR patterns of E. faecalis clinical isolates. Similarity clustering analysis was carried out using UPGMA and Dice coefficient. The dashed line is hypothetical, indicating 85% similarity. Antibiotic susceptibility, resistance determinants, and biofilm formation capacity among E. faecalis clinical isolates were reported. Antibiotic susceptibility: Black cell: Resistant, Gray cell: Intermediate, White cell: Sensitive. Biofilm formation: White cell: Biofilm Non-producer, Light gray cell: Weak Biofilm Producer, Dark gray cell: Moderate Biofilm Producer, Black cell: Strong Biofilm Producer. Resistance determinants: Black cell: Resistance gene detected, White cell: Resistance gene not detected.
Abbreviations: AM, Ampicillin; AMC, Amoxicillin/clavulanic acid; AX, Amoxicillin; AZM, Azithromycin; CIP, ciprofloxacin; CLR, Clarithromycin; DA, Clindamycin; DO, Doxycline; E, Erythromycin; HLGR, High Level Gentamicin Resistance; HLSR, High Level Streptomycin Resistance; IPM, Imipenem; LEV, Levofloxacin; LZD, Linezolid; MEM, Meropenem; ND, Not Determined; RAPD-PCR, random amplified polymorphic DNA-PCR; SAM, Ampicillin/Sulbactam; TE, Tetracycline; UPGMA, unweighted-pair group method with arithmetic averages; VA, Vancomycin.

Figure 2 Genotyping of E. faecium clinical isolates using RAPD-PCR method.
Notes: Dendrogram was created using RAPD-PCR patterns of E. faecium clinical isolates. Similarity clustering analysis was carried out using UPGMA and Dice coefficient. The dashed line is hypothetical, indicating 85% similarity. Antibiotic susceptibility, resistance determinants, and biofilm formation capacity among E. faecalis clinical isolates were reported. Antibiotic susceptibility: Black cell: Resistant, Gray cell: Intermediate, White cell: Sensitive. Biofilm formation: White cell: Biofilm Non-producer, Light gray cell: Weak Biofilm Producer, Dark gray cell: Moderate Biofilm Producer, Black cell: Strong Biofilm Producer. Resistance determinants: Black cell: Resistance gene detected, White cell: Resistance gene not detected.
Abbreviations: AM, Ampicillin; AMC, Amoxicillin/clavulanic acid; AX, Amoxicillin; AZM, Azithromycin; CIP, Ciprofloxacin; CLR, Clarithromycin; DA, Clindamycin; DO, Doxycline; E, Erythromycin; HLGR, High Level Gentamicin Resistance; HLSR, High Level Streptomycin Resistance; IPM, Imipenem; LEV, Levofloxacin; LZD, Linezolid; MEM, Meropenem; ND, Not Determined; RAPD-PCR, random amplified polymorphic DNA-PCR; SAM, Ampicillin/Sulbactam; TE, Tetracycline; UPGMA, unweighted-pair group method with arithmetic averages; VA, Vancomycin.

Figure S1 Agarose gel electrophoresis for detection of antimicrobials resistance determinants among enterococcal isolates. (A). Detection of antimicrobials resistance encoding genes among enterococcal isolates by PCR. Lane M is Molecular weight marker. 1:aac(6’)-Ie-aph(2’’)-Ia (200 bp); 2: ant(6’)-Ia (597 bp); 3: tetK (370 bp); 4: tetL (715 bp); 5: tetM (170 bp); 6: ermB (639 bp) and 7: msrA/B (400 bp); 8: optrA (1395 bp); 9: int-Tn (1046 bp); and 10: xis-Tn (194 bp) genes that are used for detection antimicrobials resistance among enterococcal isolates. (B). Multiplex PCR for detection of vancomycin resistance determinants (vanA(1030 bp),vanB(536 bp),vanC1 (822 bp), and vanC2/3 (484 bp)), E. faecium-specific (658 bp), E. faecalis-specific (941 bp) and of rrs (320 bp) genes. Lane M is Molecular weight marker. 1:E. faecalis isolate; 2:E. faecium isolate; 3:E. faecium vanB isolate; 4: E. faecalis vanC1 isolate; 5: E. faecium vanC1isolate; 6:E. faecalis vanBisolate.
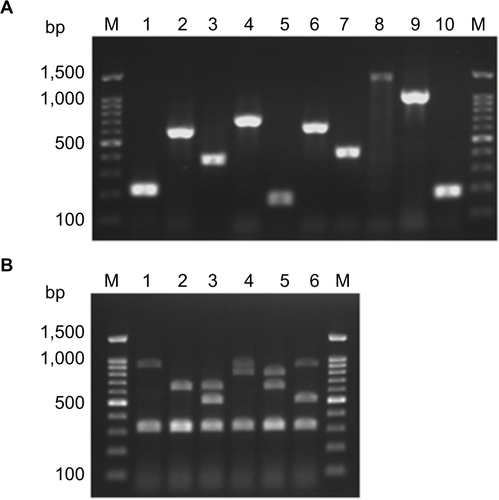
Table S1 Prevalence of antibiotic susceptibility among enterococcal species isolated from different clinical sources
Table S2 Prevalence of biofilm formation among enterococcal isolates collected from different clinical sources
Table S3 Correlation between biofilm formation capacity and antibiotic susceptibility among enterococcal clinical isolates
Table S4 Distribution of multidrug resistance (MDR) and extensive drug resistance (XDR) among enterococcal clinical isolates
