Figures & data
Figure 1 The gene of PA0847 affects biofilm formation and motility phenotypes in Pseudomonas aeruginosa. Notes: (A) Swimming motility of P. aeruginosa PAO1 (WT), the PA0847 mutants and the strains complemented with empty vector or PA0847, colony morphologies taken after 20–24 hours of incubation at 37°C, and swimming motility assays performed on medium supplemented with 0.3% agar. (B) Swarming motility of P. aeruginosa PAO1 (WT), the PA0847 mutants and the strains complemented with empty vector or PA0847, colony morphologies taken after 36–48 hours of incubation at 37°C, and swarming motility assays performed on medium supplemented with 0.45% agar. (C) Statistical analysis of biofilms of P. aeruginosa PAO1, the PA0847 mutant or the strains complemented with empty vector or PA0847. The biofilm data shown are mean ± SD of at least three replicates. PAO1: P. aeruginosa PAO1 WT; ΔPA0847-1 or ΔPA0847-2: two PA0847 transposon mutant strains; PAO1/pACYC184: PAO1 harboring the empty vector pACYC184; ΔPA0847/pACYC184: ΔPA0847-1 or ΔPA0847-2 harboring empty vector pACYC184; ΔPA0847-1/PA0847 or ΔPA0847-2/PA0847: ΔPA0847-1 or ΔPA0847-2 mutant strains harboring full-length PA0847 with its native promoter and RBS cloned into pACYC184.
Abbreviation: WT, wild type.
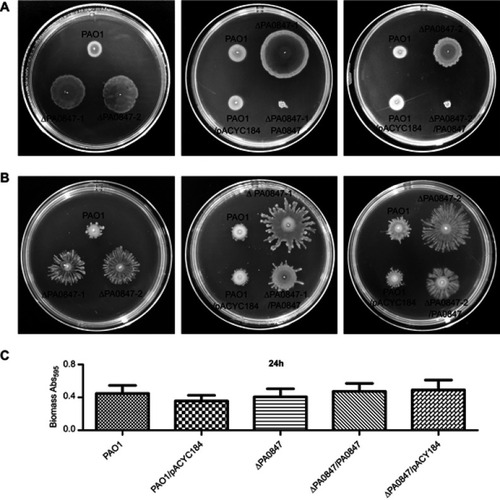
Figure 2 Pseudomonas aeruginosa PAO1 and PA0847 mutants motility and biofilm formation in response to individual amino acids differently. Notes: (A) Differential impacts of individual amino acids on bacterial motility. Swimming motility of both P. aeruginosa PAO1 and PA0847 mutants in response to five amino acids. Shown are pictures of plates incubated for 20–24 hours at 30°C. Left: wild type, upper right: ΔPA0847-1, lower right: ΔPA0847-2. (B) P. aeruginosa biofilm formation in response to individual amino acids. Shown is the quantification of 24 hour biofilms formed in the tubes and stained with Crystal Violet, solubilized in 95% ethanol and measured at 595 nm. Asterisks indicate values significantly different from the wild type. ***p<0.005.
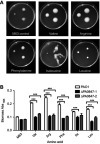
Figure 3 Diguanylate cyclase activity of PA0847 GGDEF domain. Notes: (A) Molecular architecture of PA0847. Domains are predicted by using the Conserved-Domain Database search from NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and transmembrane helices are predicted in the TMHMM Server (http://www.cbs.dtu.dk/services/TMHMM/). The amino acid number of each domain is indicated above the corresponding bar. (B) DGC activity of PA0847 GGDEF domain is detected by HPLC assays. GTP and c-di-GMP standard HPLC profiles are indicated above the assays. (C) The binding of GTPγS to PAS-GGDEF domain showed a binding affinity (Kd) of 15.3±1.3 µM. (D) Gel filtration profiles of PAS-GGDEF and GGDEF. The calculated MW of each peak based on protein standards is indicated above the peak. (E) Congo Red staining of wild-type PAO1 and ΔPA0847.
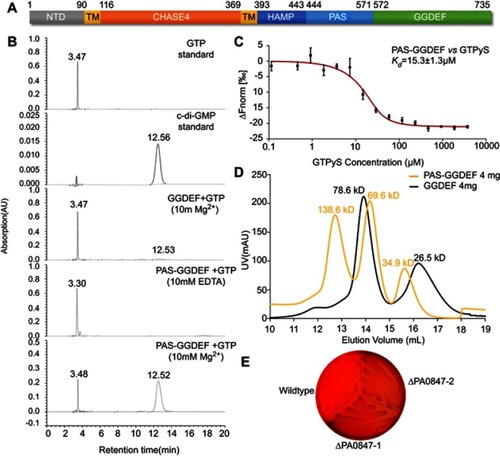
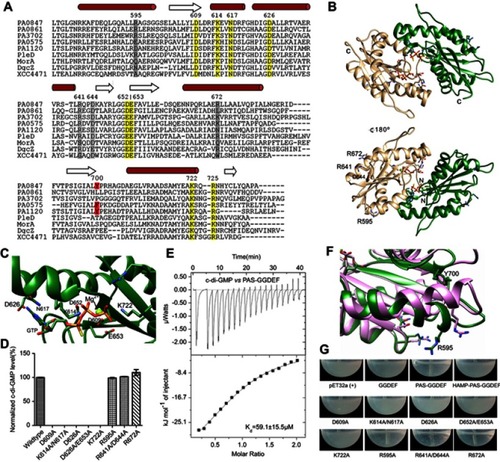
Figure 4 Structural model of PA0847 GGDEF dimer. Notes: (A) Sequence alignment between PA0847 GGDEF domain and other DGCs of known structure. Secondary structure elements are indicated above the sequences. Key residues involved in binding GTP are highlighted in yellow and putative residues in inhibition sites are highlighted in grey. (B) Structural model of PA0847 GGDEF dimer in complex with GTPs and Mg2+. (C) GTP binding residues labeled and depicted as stick models. (D) c-di-GMP synthesis by wild type and GGDEF point mutants of PA0847. (E) Binding of c-di-GMP to PAS-GGDEF dual domain. (F) Structural comparison of GGDEF domains between PA0847 (green) and PleD (pink). Bulky residue Y700 was showed in stick. (G) Different phenotype of Escherichia coli with overexpressed wild type and mutants of PAS-GGDEF domains.