Figures & data
Figure 1 Circular comparison of mcr-9-carrying IncHI2 plasmids. The complete sequence of pMCR-SCNJ07 was used as the reference. The arrows indicate deduced ORFs and their orientations. The mcr-9 gene is indicated in red. The circular maps were generated using BRIGCitation27 and plasmids were included in the following order (inner to outer circles): pMCR-SCNJ07 (this study, accession no. MK933279), pT5282-mphA (KY270852), pN1863-HI2 (MF344583), pMRVIM0813 (KP975077), pCTXM9_020038 (CP031724), pC45-VIM4 (LT991958), pSE15-SA01028 (CP026661), p505108-MDR (KY978628), pGW1 (CP028975), pRH-R27 (LN555650) and p707804-NDM (MH909331).
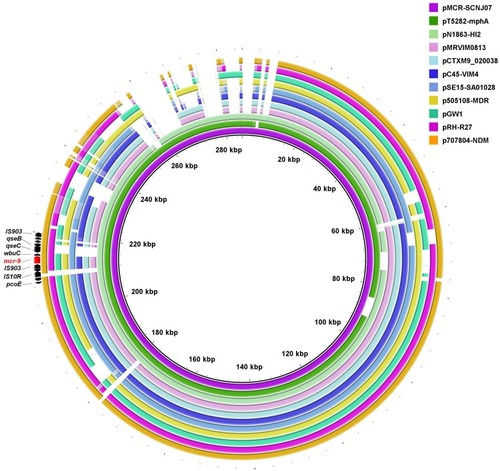
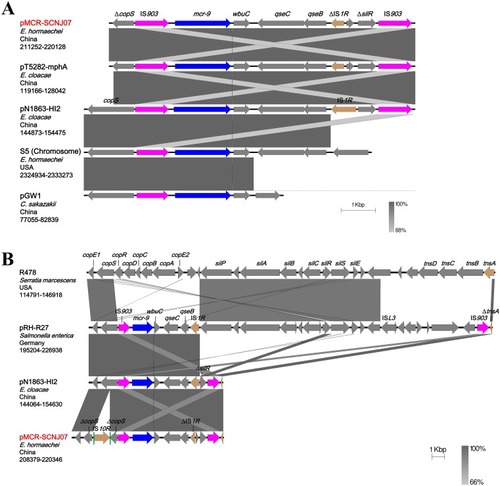
Figure 2 Colinear analyses for genetic environment of mcr-9. (A) Comparison of the mcr-9-containing regions from different plasmid reservoirs and the chromosome of E. hormaechei strain S5. (B) Comparison of the mcr-9-neighboring genetic contexts on IncHI2-type plasmids. The corresponding region on non-mcr-9-carrying plasmid R478 (top) is shown for comparison. Arrows indicate the positions and directions of the genes and Δ indicates the truncated gene. mcr-9 genes are indicated in dark blue and IS903 are highlighted in purple. Gray shades denote shared regions with a high degree of homology. Vertical green bars represent the direct repeats of IS10R. The accession numbers were: E. hormaechei strain S5 (Accession no. CP031571), pT5282-mphA (KY270852), pN1863-HI2 (MF344583), pGW1 (CP028975), R478 (NC_005211), pRH-R27 (LN555650), pN1863-HI2 (MF344583), and pMCR-SCNJ07 (this study, MK933279). The construction of sequence comparison was performed using BLAST (http://blast.ncbi.nlm.nih.gov) and Easyfig version 2.2.3.Citation26