Figures & data

Table 1 Summary of Some Clinical Studies with IA Methods
Table 2 Commercial Diagnostic Kits, Platforms and Systems Using IA Techniques
Figure 1 Schematic illustration of LAMP. The reaction begins with annealing of the FIP and its extension by polymerase enzyme to form a new strand of DNA which is displaced by extension of F3 (non-cyclic phase). The new strand forms a loop at the 5ʹ end with compliment sequences at F1 and F1c. BIP also anneals, and a similar process as of FIP releases another new strand that forms a double stem-loop structure at both ends. A new double stem-loop DNA with a sequence complementary to the first strand and another stem-loop with double the length of the previous one is formed subsequently from self-primed extension by the DNA strand, and by annealing of FIP and BIP coupled with extensions and displacements. As products from subsequent steps get involved in the reaction, DNA structures of different sizes bearing inverted repeats on the same strand result with multiple loops formed as a result of alternately repeated sequences on the same strand. Reproduced from Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63–E63.Citation16
Figure 2 Helicase-dependent amplification. The process begins with strand separation by helicase, primer annealing, extension by polymerase and ends with two copies of the target. Adapted from Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. Copyright © 2004 European Molecular Biology Organization.Citation56
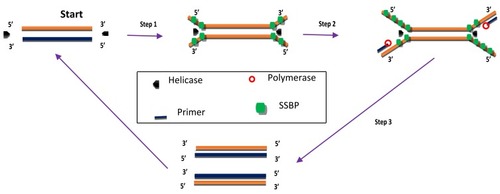
Figure 3 Signal-mediated amplification of RNA technology. A 3WJ forms; DNA polymerase extends the extension probe; T7 RNA polymerase promoter is formed, and T7 RNA polymerase generates signals (1st RNA). Signals can be transcribed by T7 RNA polymerase and detected by ELOSA (option 2) or further amplified to yield multiple 2nd RNA signals (option 1) which can then be detected. Adapted with permission from Springer Nature. Wharam SD, Hall MJ, Wilson WH. Detection of virus mRNA within infected host cells using an isothermal nucleic acid amplification assay: marine cyanophage gene expression within Synechococcus sp. Virol J. 2007;4(1):52.Citation68
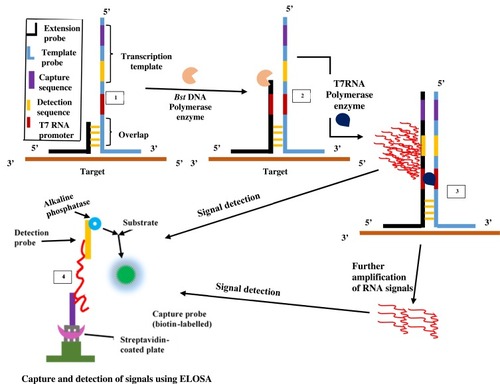
Figure 4 Nucleic acid sequence-based amplification. P1 anneals to sense RNA, then extension by AMV reverse transcriptase enzyme follows, forming a hybrid of RNA and complementary DNA (cDNA). P2 binds to cDNA following RNA degradation by RNase H; extension by reverse transcriptase generates DNA with T7 RNA polymerase promoter sequence, which transcribes into multiple antisense RNA (-) copies by DNA dependent RNA polymerase. P2 anneals to resulting RNA (-) strands, undergoes extension and forms hybrids of RNA (-) and cDNA. RNA (-) is degraded again by RNase H while P1 binds to the new cDNA, followed by extension by reverse transcriptase to yield dsDNA bearing promoter sequence for T7 RNA polymerase and the cycle continues, generating multiple RNA copies. Adapted from Zanoli LM, Spoto G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors. 2012;3(1):18–43.Citation59
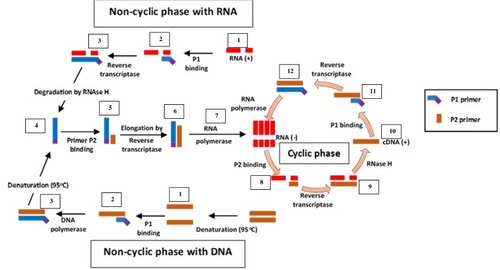
Figure 5 Recombinase polymerase amplification. Recombinase-bound primers scan for complementary sequence; recombinase enzyme separates the strands and SSB proteins prevent reannealing of separated strands; primers anneal, and extension by DNA polymerase takes place, leading to the formation of two dsDNA. Repetition of this cycle produces multiple copies of DNA. Adapted from Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204–e204.Citation93
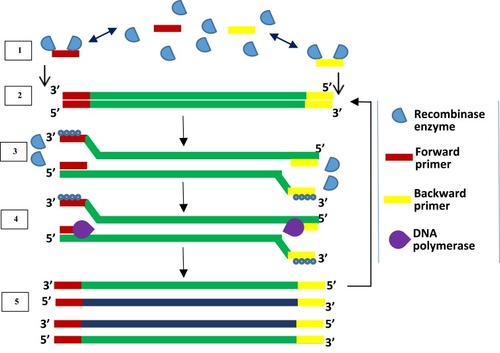
Figure 6 Rolling circle amplification. [1] Primer anneals to circular DNA which is continuously extended by polymerase enzyme to produce a displaced long ssDNA with tandem repeat sequences. Adapted with permission from Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J Am Chem Soc.1996;118(7):1587–1594. Copyright 1996 American Chemical Society.Citation203 [2] Hyper-branched RCA in which more than one primer anneals to products of previous amplification or padlock probe is used to form a circular DNA. New strands arise from the extension of primers; as they are displaced, reverse primers anneal and are extended to generate more displaced strands, resulting in multiple copies of highly branched DNA. Adapted from Yan J, Zhou Y, Zheng AS, et al. Isothermal amplified detection of DNA and RNA. Mol. BioSyst. 2014;10:970-1003.Citation70 [3] Multiprimed RCA or multiply primed RCA in which multiple primers anneal to different points on a circular DNA with simultaneous extension and displacement. Multiply-branched structures result. Adapted with permission from Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11(6):1095–1099.Citation134 Copyright © 2001, Cold Spring Harbor Laboratory Press.
![Figure 6 Rolling circle amplification. [1] Primer anneals to circular DNA which is continuously extended by polymerase enzyme to produce a displaced long ssDNA with tandem repeat sequences. Adapted with permission from Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J Am Chem Soc.1996;118(7):1587–1594. Copyright 1996 American Chemical Society.Citation203 [2] Hyper-branched RCA in which more than one primer anneals to products of previous amplification or padlock probe is used to form a circular DNA. New strands arise from the extension of primers; as they are displaced, reverse primers anneal and are extended to generate more displaced strands, resulting in multiple copies of highly branched DNA. Adapted from Yan J, Zhou Y, Zheng AS, et al. Isothermal amplified detection of DNA and RNA. Mol. BioSyst. 2014;10:970-1003.Citation70 [3] Multiprimed RCA or multiply primed RCA in which multiple primers anneal to different points on a circular DNA with simultaneous extension and displacement. Multiply-branched structures result. Adapted with permission from Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11(6):1095–1099.Citation134 Copyright © 2001, Cold Spring Harbor Laboratory Press.](/cms/asset/c8df30bf-9cda-4a30-b7b3-1fa38215905a/didr_a_217571_f0006_c.jpg)
Figure 7 Strand displacement amplification. Denaturation of a dsDNA allows annealing of primers S1, S2, B1 and B2 which are extended simultaneously with the displacement of extended S1 and S2 strands bearing HincII restriction sites. When a dsDNA with both strands bearing HincII restriction sites is generated, the unmodified original strand extended from the primers is nicked by the enzyme, while the polymerase extension of the 3ʹ end of the newly synthesized strand results in the displacement of downstream DNA strands. The displaced strands then form the template for subsequent amplification which results in exponential amplification of the target. Adapted with permission from Walker TG. Empirical Aspects of Strand Displacement Amplification. Genome Res. 1993; 3:1–6.Citation204 Copyright © 1993, Cold Spring Harbor Laboratory Press.
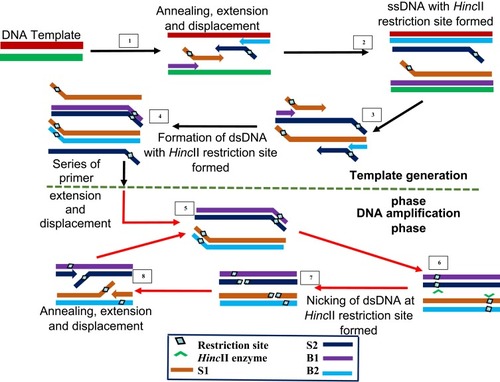
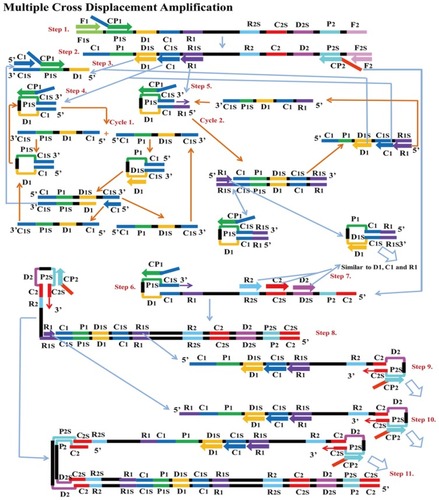
Figure 8 Multiple cross displacement amplification. CP1 anneals to P1s to begin the reaction. Synthesis by F1 displaces the product formed by CP1, which is then annealed to by amplification primers D1, C1, R1, CP2 (cross primer) and F2 (displacement primer), leading to four different products (step 2). A new product CP1/D1 is formed by the extension of product D1 by C1 and CP1 (step 3), while the C1 product forms an intramolecular stem loop due to complementarity at its 3ʹ (C1s sequence) and 5ʹ (C1 sequence) ends. CP1 extends the stem loop, forming a new CP1/C1 product (step 4). These can then become templates from which amplification primers D1, C1 and CP1 generate products CP1/D1 and CP1/C1 respectively as before (Cycle 1). In cycle 2, product R1 undergoes a similar amplification process as C1 to yield products CP1/C1, CP1/D1, CP1/R1 and R1/R1s respectively. CP2 product displaced by F2 primer from step 2, is extended at the 3ʹ end to form a new double strand with stem loop structure (step 6). Primers C2, D2 and R2 then anneal to the newly formed strands to repeat similar form of amplification by C1, D1 and R1 (step 8). The two cycles generate several stem loop structures which become templates for exponential amplification to yield multiple copies of the target DNA with numerous inverted repeats. Reproduced with permission from Wang Y, Wang Y, Ma A-J, et al. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci Rep. 2015;5:11902.Citation25