Figures & data
Table 1 The Sequence Of Primers Of The Genes Used In This Study
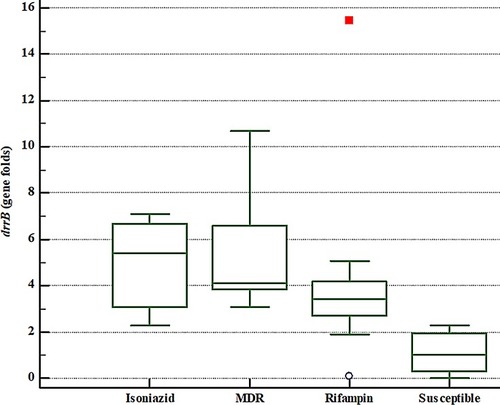
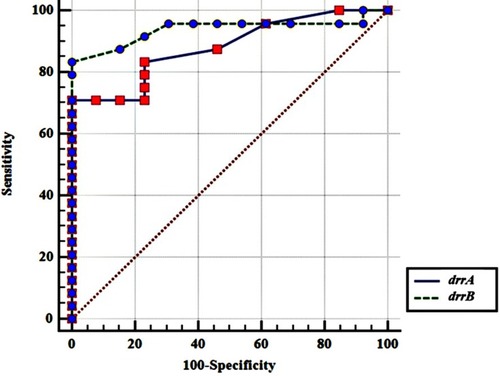
Table 2 The Results From Phenotypic Drug Susceptibility Testing And Relevant drrA And drrB Gene Expression Fold Changes For MTB Isolates
Table 3 Correlation Of Genes Expression Fold Changes Of drrA And drrB In Mycobacterium Tuberculosis Isolates With Various Drug-Resistance Patterns
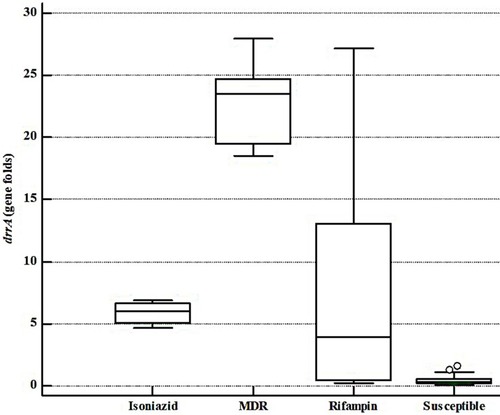
Figure 1 The drrA gene expression fold comparison of different groups of MTB isolates as per DST results.