Figures & data
Table 1 Baseline Characteristics of Patients and Antimicrobial Treatments
Figure 1 Colony morphology of the A. baumannii isolates on MH plates. Isolate 403 was mucoid with moist, irregular round colonies, and isolate 404 was matt with round and neat edge colonies.
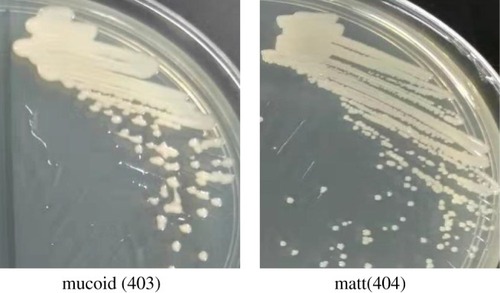
Figure 2 Capsule thickness. (A-D) (A) Matt isolate 404; (B) mucoid isolate N25; (C, D) mucoid isolate 405 and matt isolate 406, isolated from the same patient. (E) The average capsule thickness of different capsular types strains. Mucoid A baumannii isolate was more than 2.5 times that of the matt phenotype (p < 0.001); (F) Capsule thickness of 8 isolates, isolated from 4 patients, one matt isolate and one mucoid isolate were isolated from each patient. *p < 0.05; ***p < 0.0001.
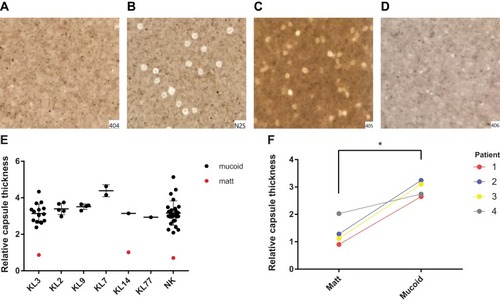
Table 2 Antimicrobial Susceptibility in A. Baumannii
Table 3 adeN and adeR Mutation Conferred Tigecycline Resistance
Figure 3 Minimum spanning tree based on cgMLST analysis. Each circle represents an allelic profile, i.e. genotype, based on sequence analysis of up to 2390 target genes. The numbers on the connecting lines illustrate the numbers of target genes with different alleles.
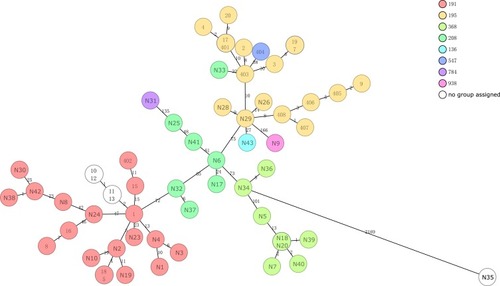
Figure 4 Resistance genes. The isolate N35 only had two resistance genes, which was susceptible to all 11 antibiotics (). The isolate N32 and N37 contained four resistance genes, and the remaining 57 isolates all contained more than five resistance genes.
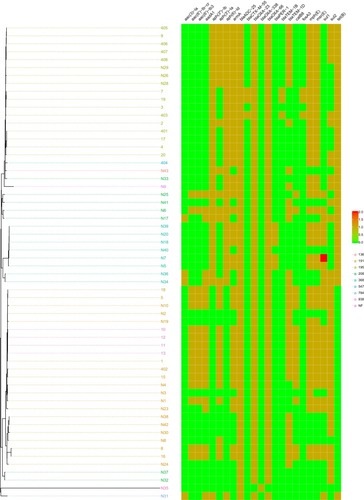
Table 4 SNPs Analysis of Isolate 405 (Mucoid) and 406 (Matt)
Table 5 SNPs Analysis of Isolate 407 (Mucoid) and 408 (Matt)
Figure 5 Biofilm-formating ability and relative growth rate. (A) Biofilm-forming ability of different capsular types. (B) Relative growth rate of different capsular type strains, taking the isolate 1 as reference. Both biofilm-forming ability and relative growth rate had no significant differences between matt and mucoid isolates.
Abbreviation: NK, unknown capsular types.
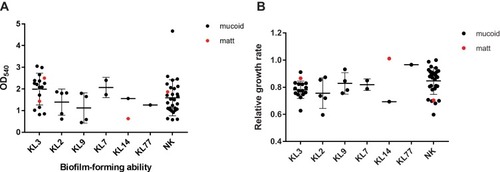
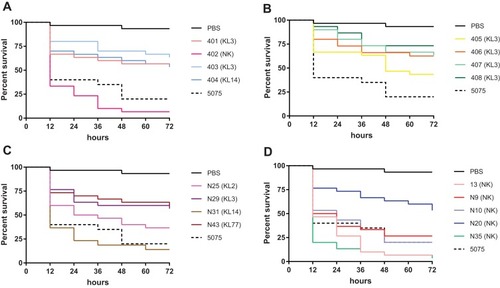
Figure 6 Percent survival of G. mellonella infected with A. baumannii. (A) 401 (mucoid, ST195, KL3) and 402 (matt, ST191, NK) isolated from one patient, and (mucoid, ST195, KL3) and 404 (matt, ST547, KL14) isolated from one patient; (B) 403 (mucoid) and 404 (matt) isolated from one patient, and (mucoid) and 404 (matt) isolated from one patient, all 4 isolates belong to ST195, KL3; (C, D) all isolates were mucoid.
Abbreviation: NK, unknown capsular types.