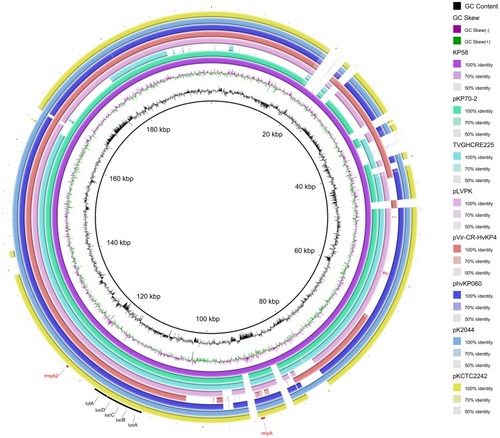
Figures & data
Figure 1 Recombination-filtered core genome phylogeny and the distribution of antimicrobial resistance genes for ST11 K. pneumoniae isolates recovered from Hangzhou, China. The cell in different colour indicates the presence of the gene while the blank cell indicates the absence of the gene.
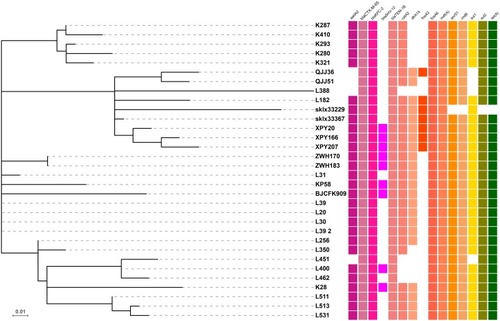
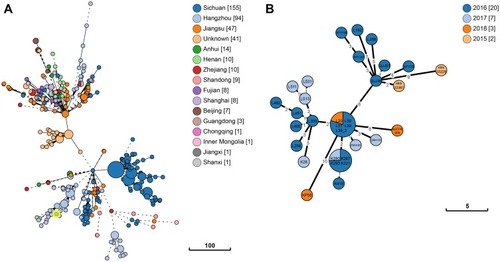
Figure 2 Phylogenetic relationship between K. pneumoniae KP58 and closely related K. pneumoniae strains presently retrieved from BacWGSTdb. Clonal relationships between different isolates are depicted by the line length connecting each circle. The numbers given in square brackets indicate the number of isolates from each country. (A) The minimum spanning tree of all ST11 K. pneumoniae strains recovered from China. (B) The minimum spanning tree of the closely related strains with K. pneumoniae KP58 (differing by <10 cgMLST loci).