Figures & data
Table 1 MIC Distributions for Various Mycobacteria Species Against PA-824
Figure 1 Multiple sequence alignments and phylogenetic analysis of Ddn sequences in MTB and its homologues of NTMs. (A) Multiple sequence alignments of Ddn of MTB and its homologues of NTMs. Shared amino acids are highlighted in red and stars mark non-shared amino acids selected for testing in this study. (B) Phylogenetic tree of MTB Ddn and NTMs Ddn homologous sequences constructed based on protein sequence similarity.
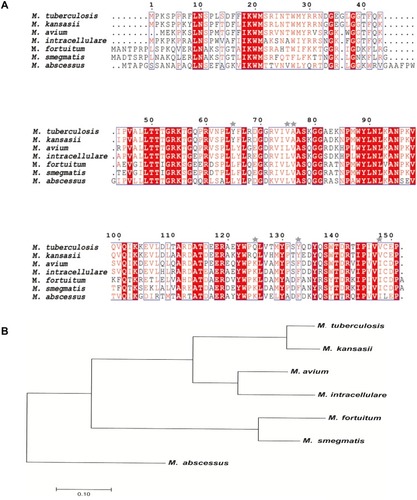
Figure 2 Protein expression analysis and growth curve analysis of M. smegmatis transformed with plasmids encoding MTB Ddn and its mutants. (A) Western blot analysis of MTB Ddn and Ddn mutants recombinantly expressed by M. smegmatis. (B) Growth curve analysis of M. smegmatis transformed with plasmids encoding MTB Ddn and Ddn mutants.
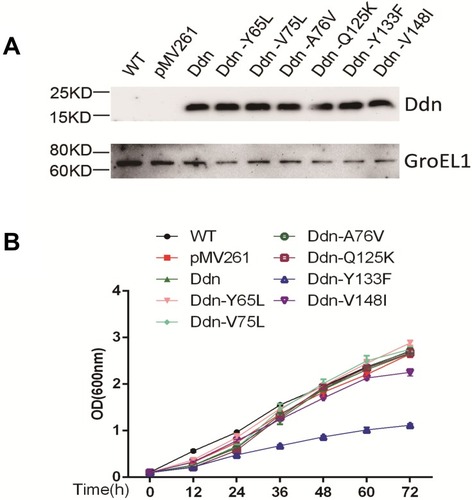
Table 2 MIC of Ddn Mutants in M. smegmatis Against PA-824
