Figures & data
Figure 1 Potential genome manipulation using ZFNs, TALENs, and Cas9. ZFNs, TALENs, and Cas9 can be designed to target any gene in the genome of prokaryotic and eukaryotic cells. They are delivered to the cells via transduction or electroporation. Each ZFNs and TALENs contain non-specific DNA-cleaving domain (FokI), and DNA-binding domain. Each DNA-binding domain of ZFNs and TALENs recognizes 3–4 and 1 DNA sequence, respectively. Each repeat of TALENs is 33–35 amino acids in length, with two RVD (NI=A, NG=T, HD=C, and NK=G). The spacer regions between the monomers of TALENs and ZFNs are 6–40 bp and 5–7 bp in length, respectively. Dimerization of FokI is necessary for DNA cleavages within the spacer regions between the two bindings. In Cas9, gRNA recognizes and bind to targeted sequence, and Cas9 cleaves double-strand DNA in 3 bp upstream of the PAM sequence (NGG). The double-strand break can stimulate the natural DNA-repair mechanisms of the cell including nonhomologous end joining repair and homologous repair, followed by can be harnessed to create gene knock-out or knock-in. In addition, nonhomologous integration and repair is trended in biotechnology.
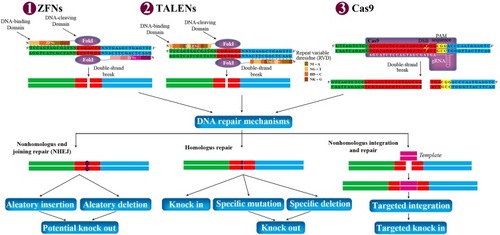
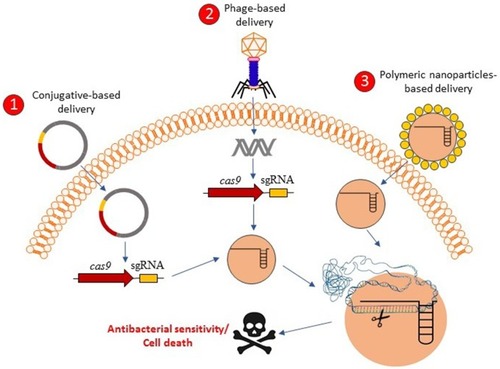
Figure 2 Graphical concepts model of CRISPR-Cas delivery for antibacterial affecting. CRISPR-Cas-based antibacterials could be delivered into the bacterial cells through the three proposed delivery mechanisms including 1) conjugative-based delivery, 2) phage-based delivery, and 3) polymeric-nanoparticles-based delivery. After the delivery of CRISPR-Cas systems in the bacterial cells, the bacterial cells might be resensitized against antibacterial agents or be killed based on the antibacterial resistant genes targets or essential genes targets, respectively.